Research Article, J Appl Bioinforma Comput Biol Vol: 9 Issue: 3
A Bioinformatics Study of SARS-CoV-2 Surface Glycoprotein in Indian Perspective
Anindita Dey1,2*, Sumanta Dey2,3 and Papiya Nandy2
Department of Biostatistics, Epidemiology and Environmental Health Sciences, Jiann-Ping Hsu College of Public Health, Georgia Southern University, Statesboro, GA 30460, USA
*Corresponding Author: Anindita Dey
Deprtment of Botany, Asutosh College, Kolkata-26, Centre for Interdisciplinary Research and Education, 404 B Jodhpur Park, Kolkata-68, India
Tel: 8240737707
E-mail: sunimondal@yahoo.com
Received: July 11, 2020 Accepted: July 22, 2020 Published: July 29, 2020
Citation: Dey A, Dey S, Nandy P (2020) A Bioinformatics Study of SARS-Cov-2 Surface Glycoprotein in Indian Perspective. J Appl Bioinforma Comput Biol 9:2. doi: 10.37532/jabcb.2020.9(2).169
Abstract
The 20th-century world is facing its biggest problem to combat with deadly SARS- CoV-2, a virus that has locked us down in a new world and 5,47,321 lives have already been sacrificed. The only possible way to cope with this virus is to design a suitable drug or a vaccine. India at present is also on the same platform with ~ 7, 46,500 positive cases and a death toll of 20,684. Since the viral spike protein is the outermost surface-exposed protein and responsible for viral entry into the host cell, it is important to characterize the spike protein for the development of any therapeutic response to infections from this virus. In this article we done the in-silico analysis of the Indian spike protein by studying phylogenetic relationship, nucleotide and amino acid characterization, codon usage bias, transition/transversion matrix, hydropathy index, parameters for protein characterization and epitope prediction in different aspects. Our further analysis shows some reasonable potential epitope regions which would be effective for vaccine designing and can elicit an immune response against the viral infection.
Keywords: SARS-CoV-2; Spike protein; phylogenetic tree; 2D graphical representations; transition/transversion ratios; amino acid composition; hydropathy profile; codon usage bias; epitopes; pathogenicity; vaccines
Introduction
The world threatened by the pandemic disease ‘corona’ announced by the World Health Organisation (WHO) on 11th March 2020, is caused by SARS-CoV-2, the well-known Novel Corona Virus which was first isolated in Wuhan city of China [1]. The existence of corona virus was first identified in the mid of 1960 causing infection to varieties of animals as well as human beings [2]. Severe acute respiratory syndrome (SARS-CoV) was quite common from 2002 and Middle East Respiratory Corona Virus (MARS-CoV) was known to us since 2012 [3-6]. However, in 2020, the most serious and deadly human pathogen, SARS-CoV-2 develops a global challenge to prevent the virus as early as possible to rescue the human population. On 30th January 2020, WHO declared this as a public health emergency of international concern (PHEIC). People attacked by this virus are suffering from one or more symptoms like nasal congestion, headache, runny nose, conjunctivitis, sore throat, diarrhoea, loss of taste, smell, a rash on skin or discoloration of fingers or toes and mild to heavier respiratory problems. The coronavirus is a singlestranded RNA virus that belongs to the family Coronaviridae under the order Nidovirales and the genus is Betacoronavirus [7-9]. Among all RNA viruses this virus possesses 26.4 to 31.7 kb genomes that are the largest one which is enclosed within a capsid made up of matrix protein.
Coronavirus membrane possesses three to four types of proteins comprising of membrane protein (M), spike proteins (S), nucleocapsid (N) and envelope protein (E) among which the most abundant one is M. The SARS-CoV-2 enters within the host cell through its homotrimeric, transmembrane spike glycoprotein (S), protrudes from the viral surface having a crown like appearance [10]. The spike protein comprises of two functional subunits-S1 and S2. The S1 subunit helps to bind with the host cell receptor and S2 plays a role during the fusion of viral and cellular membranes. There is a unique N-terminal fragment within the spike protein of the viral genome leaving a short -NH2-terminal domain outside the virus and a long -COOH terminus inside the virion [11-16].
The understanding of the viral spike protein is very much necessary to produce drugs as well as vaccine design. So, in the present scenario, the detailed characterization of the surface exposed spike protein of the deadly pathogenic virus has the prime importance to the researchers. In this article we have done the utmost characterization of Indian spike glycoprotein of SARS-CoV-2 covering its phylogenetic relationship, nucleotide and amino acid characterization, codon usage bias, transition/transversion matrix, hydropathy index, parameters for protein characterization and epitope prediction.
Materials and Methods
Sequence data of ten Indian Spike glycoprotein genes for SARSCoV- 2 were downloaded in random basis from the NCBI GenBank database. The list of the sequence IDs is given in Table 1.
| locus_id | Amino acid length | Source | Collection date | Product |
|---|---|---|---|---|
| QHS34546 | 1272 | "India: Kerala State" | "2020-01-27" | "surface glycoprotein" |
| QIA98583 | 1273 | "India: Kerala State" | "2020-01-31" | "surface glycoprotein" |
| QJC19491 | 1273 | "India: Rajkot" | "2020-04-05" | "surface glycoprotein" |
| QJF11812 | 1273 | "India" | "2020-04-08" | "surface glycoprotein" |
| QJF11824 | 1273 | "India" | "2020-04-08" | "surface glycoprotein" |
| QJF11836 | 1273 | "India" | "2020-04-14" | "surface glycoprotein" |
| QJF11848 | 1273 | "India" | "2020-04-10" | "surface glycoprotein" |
| QJF11860 | 1273 | "India" | "2020-04-10" | "surface glycoprotein" |
| QJF11872 | 1273 | "India" | "2020-04-06" | "surface glycoprotein" |
| QJF11884 | 1273 | "India" | "2020-04-14" | "surface glycoprotein" |
Table 1: List of Indian Spike glycoprotein sequences downloaded from NCBI.
The genes are plotted in a 2D graphical representation scheme [17] and analyzed to see the base distribution pattern [18]. The sequences are plotted as a walk in a 2D grid taking one step in the negative x-direction for an adenine, in the positive y-direction for guanine, the positive x-direction for a thymine/uracil and in the negative y-direction for a cytosine. This yields (x,y) co-ordinates for each base in a sequence, successive plots of the points yields a curve in the 2D graph [17]. We also did an alignment based matrix to draw phylogenetic tree in order to find the evolutionary trend between the Indian spike sequences with other sequences from effected countries of the world by using the software MEGA 5.2 [19]. The transition/ transversion ratio along with the amino acid composition and codon usage bias of the Spike gene sequences were also computed using the same software. To understand the protein statistics in more details we use PEPSTATS [20-21]. IEDB server and ABCpred were used to search the T-cell and B-cell epitope regions in the Spike protein structure [22-23].
Result
The phylogenetic relationship between the genome of the SARS CoV-2 strains, taking sequences from each largely effecting country randomly, is displayed in Figure 1. The result shows that the Indian strain belongs to a clade with other Asian strains that includes China, Vietnam, Nepal along with Wuhan while the American and the European strains remain in a separate clade. However, South Korean strain represents a single clade.
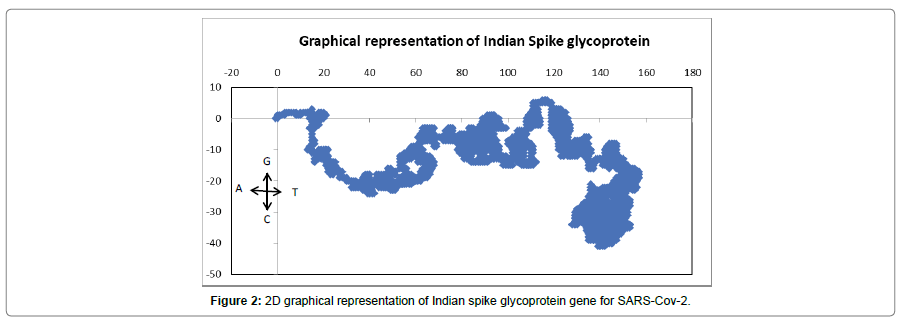
To get a more detail view of the base distribution of the Indian spike glycoprotein gene, the nucleotide sequences are plotted in a 2D graphical representation scheme described in the method and material section described above. The 2D plot (Figure 2) shows more thymine and cytosine amount in the gene base pair composition which implies more pyrimidine bases than purine.
The data of Table II shows the G-A/C-G ratio for Indian SARS-CoV-2 is 14.74 which imply the higher transition rate than transversion. However, according to Duchene (2015) mammalian genes have a transition-transversion ratio of 2 to 5 approximately while RNA viruses with their mutation rate about 15 or 20 [24]; which also reflects in our previous study in case of Flaviviruses [25] compare to Indian SARS-Cov-2.
| From\To | A | T | C | G |
|---|---|---|---|---|
| A | - | 3.9238 | 2.2294 | 20.0134 |
| T | 3.4694 | - | 8.8453 | 2.1707 |
| C | 3.4694 | 15.5681 | - | 2.1707 |
| G | 31.9866 | 3.9238 | 2.2294 | - |
Table 2: Transition-transversion rate matrix of different strains of SARS-CoV-2 for Indian spike glycoprotein genes.
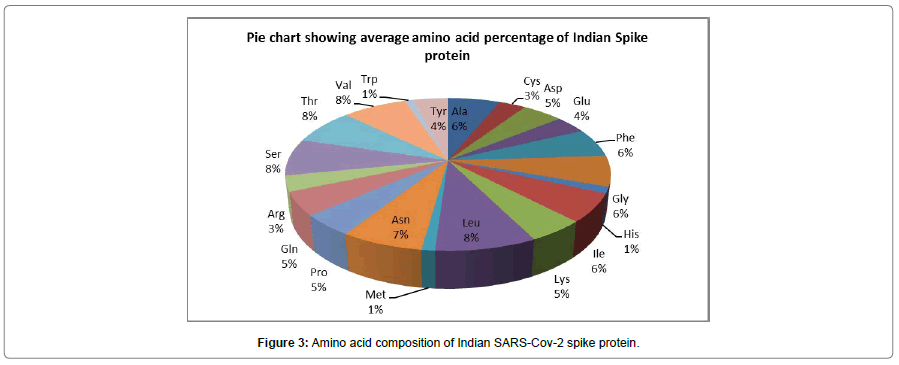
Fig III shows the different amino acid composition of spike glycoprotein for Indian SARS-CoV-2. The result implies higher percentage of hydrophobic amino acids like Leucine, Valine, Alanine, Phenylalanine and hydrophilic amino acids like Asparagine, Glutamine, Aspartic acid and Serine. Different statistics like molecular weight, isoelectric point, aliphatic, aromatic, polar, non-polar with basic and acidic properties for individual protein sequence are shown in the Table III.
| Protein IDs | Molecular weight | Isoelectric Point | Aliphatic | Aromatic | Non-polar | Polar | Charged | Basic | Acidic |
|---|---|---|---|---|---|---|---|---|---|
| QHS34546 | 140972.27 | 6.5293 | 28.381 | 12.5 | 54.796 | 45.204 | 18.003 | 9.355 | 8.648 |
| QIA98583 | 141206.52 | 6.6146 | 28.28 | 12.569 | 54.753 | 45.247 | 18.068 | 9.427 | 8.641 |
| QJC19491 | 141148.49 | 6.7881 | 28.28 | 12.569 | 54.831 | 45.169 | 18.068 | 9.505 | 8.562 |
| QJF11812 | 141164.47 | 6.7019 | 28.28 | 12.647 | 54.831 | 45.169 | 17.989 | 9.427 | 8.562 |
| QJF11824 | 141120.43 | 6.7006 | 28.28 | 12.569 | 54.831 | 45.169 | 17.989 | 9.427 | 8.562 |
| QJF11836 | 141178.47 | 6.6146 | 28.28 | 12.569 | 54.753 | 45.247 | 18.068 | 9.427 | 8.641 |
| QJF11848 | 141178.47 | 6.6146 | 28.28 | 12.569 | 54.753 | 45.247 | 18.068 | 9.427 | 8.641 |
| QJF11860 | 141178.47 | 6.6146 | 28.28 | 12.569 | 54.753 | 45.247 | 18.068 | 9.427 | 8.641 |
| QJF11872 | 141120.43 | 6.7006 | 28.28 | 12.569 | 54.831 | 45.169 | 17.989 | 9.427 | 8.562 |
| QJF11884 | 141178.47 | 6.6146 | 28.28 | 12.569 | 54.753 | 45.247 | 18.068 | 9.427 | 8.641 |
Table 3: Various statistics on the protein properties for Indian Spike Glycoprotein.
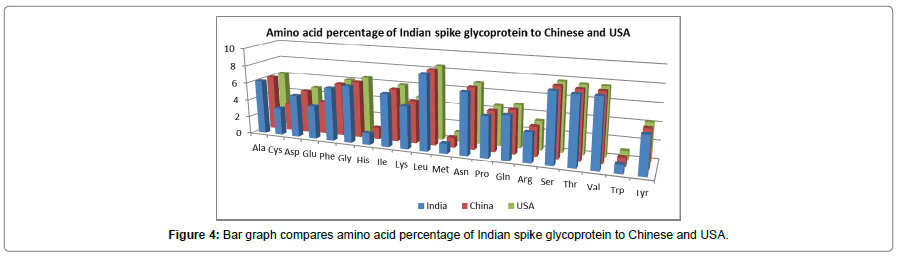
Then we compare the amino acid percentage of Indian spike glycoprotein with China and USA, since China being the first epicenter and USA hits the highest toll in term of infection as well as mortality till now. The result (Figure 4) shows almost unaltered amino acid composition between the three countries which indicates lower mutation rate at the spike protein level. These findings definitely show promising insights for researchers trying to create an effective vaccine against the virus.
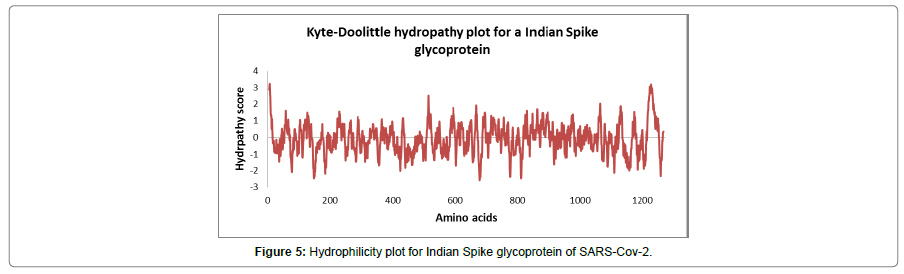
The figure (Figure 5) shows the Kyte-Doolittle hydrophilicity plot for Indian Spike glycoprotein of SARS-CoV-2 which is a quantitative analysis of the degree of hydrophobicity or hydrophilicity of amino acids of a protein. It is used to characterize or identify possible structures or domains of a protein. The plot has an amino acid sequence of the glycoprotein on its x-axis and degree of hydrophobicity and hydrophilicity on its y-axis. The amino acids at the end of the protein show higher average hydropathy.
The codon usage bias report (Table 4) shows considerable differences in codon usage. This suggests that qualitative changes have taken place between the sequences and perhaps a consequence of synonymous or non- synonymous mutations [26].
| Codon | Count | Codon | Count | Codon | Count | Codon | Count |
|---|---|---|---|---|---|---|---|
| UUU(F) | 59.1 | UCU(S) | 37 | UAU(Y) | 40.5 | UGU(C) | 27.9 |
| UUC(F) | 18 | UCC(S) | 12 | UAC(Y) | 13.5 | UGC(C) | 12 |
| UUA(L) | 28 | UCA(S) | 26 | UAA(*) | 1 | UGA(*) | 0 |
| UUG(L) | 20 | UCG(S) | 2 | UAG(*) | 0 | UGG(W) | 12 |
| CUU(L) | 36 | CCU(P) | 29 | CAU(H) | 13 | CGU(R) | 9 |
| CUC(L) | 12 | CCC(P) | 4 | CAC(H) | 4 | CGC(R) | 1 |
| CUA(L) | 9 | CCA(P) | 25 | CAA(Q) | 45.9 | CGA(R) | 0.1 |
| CUG(L) | 3 | CCG(P) | 0 | CAG(Q) | 16 | CGG(R) | 2 |
| AUU(I) | 44 | ACU(T) | 44 | AAU(N) | 54 | AGU(S) | 17 |
| AUC(I) | 14 | ACC(T) | 10 | AAC(N) | 34 | AGC(S) | 5 |
| AUA(I) | 18 | ACA(T) | 40 | AAA(K) | 38 | AGA(R) | 20 |
| AUG(M) | 14 | ACG(T) | 3 | AAG(K) | 23 | AGG(R) | 10 |
| GUU(V) | 48 | GCU(A) | 42 | GAU(D) | 42.5 | GGU(G) | 47.5 |
| GUC(V) | 21 | GCC(A) | 8 | GAC(D) | 19 | GGC(G) | 15 |
| GUA(V) | 15 | GCA(A) | 27 | GAA(E) | 34 | GGA(G) | 17 |
| GUG(V) | 13 | GCG(A) | 2 | GAG(E) | 14 | GGG(G) | 3 |
Table 4: The codon usage count for Indian Spike glycoprotein of SARS-CoV-2.
The next step was to determine the epitope regions for the Spike glycoprotein so that we can use those peptide segments from the spike protein as epitopes within the human host by adding suitable adjuvants to enhance the immune response in humans for vaccine design [27].
Our interest being the generation of antibody response to the invading pathogens, we concentrate on MHC Class II molecules that mediate the establishment of humoral immunity [28-35].
For epitope prediction we use the Immune Epitope Database and Analysis Resource (IEDB) server to determine the binding affinities for Human Leukocyte Antigens (HLA) mainly for MHC Class II for T-cells. The HLA alleles were chosen to provide coverage of around 90% of the target population which in this case was India. All predictions were done using the IEDB consensus method. The list of the binding affinities for MHC Class II T-cell epitopes, with percentile rank where low rank implies higher binding affinity, a percentile rank of 10%, below are considered good binding strength. The default peptide length used by IEDB for binding strength computations is 15 residues. Here, we give the best epitope region for individual HLA alleles shown in Tables 5 and 6.
| Allele | Start | end | Peptide | percentile_rank |
|---|---|---|---|---|
| HLA-DRB1*01:01 | 511 | 525 | VVLSFELLHAPATVC | 0.03 |
| HLA-DRB1*08:01 | 627 | 641 | DQLTPTWRVYSTGSN | 0.15 |
| HLA-DRB1*11:01 | 447 | 461 | GNYNYLYRLFRKSNL | 0.22 |
| HLA-DRB1*15:02 | 447 | 461 | GNYNYLYRLFRKSNL | 0.24 |
| HLA-DRB1*09:01 | 884 | 898 | SGWTFGAGAALQIPF | 0.33 |
| HLA-DRB1*07:01 | 713 | 727 | AIPTNFTISVTTEIL | 0.4 |
| HLA-DRB1*03:01 | 208 | 222 | TPINLVRDLPQGFSA | 0.59 |
| HLA-DRB1*04:01 | 959 | 973 | LNTLVKQLSSNFGAI | 0.82 |
| HLA-DRB1*12:01 | 957 | 971 | QALNTLVKQLSSNFG | 1.23 |
Table 5: IEDB prediction of binding affinity for MHC II of allele HLA-DRB.
| Allele | start | End | Peptide | percentile_rank |
|---|---|---|---|---|
| HLA-DQA1*05:01/DQB1*03:01 | 1216 | 1230 | IWLGFIAGLIAIVMV | 0.51 |
| HLA-DQA1*01:01/DQB1*05:01 | 483 | 497 | VEGFNCYFPLQSYGF | 1.4 |
| HLA-DQA1*05:01/DQB1*02:01 | 617 | 631 | CTEVPVAIHADQLTP | 2.9 |
| HLA-DPA1*01:03/DPB1*02:01 | 504 | 518 | GYQPYRVVVLSFELL | 0.36 |
Table 6: IEDB prediction of binding affinity for MHC II of allele HLA-DP/DQ.
We also determined the B-cell epitopes for antibody using the ABCpred server. The best possible peptides up to rank 10 are given in Table VII.
| Rank | Sequence | Start position | Score |
|---|---|---|---|
| 1 | AGTITSGWTFGAGAAL | 879 | 0.97 |
| 2 | GVSVITPGTNTSNQVA | 594 | 0.95 |
| 2 | GWTAGAAAYYVGYLQP | 257 | 0.95 |
| 2 | PQIITTDNTFVSGNCD | 1112 | 0.95 |
| 3 | HRSYLTPGDSSSGWTA | 245 | 0.92 |
| 3 | QKEIDRLNEVAKNLNE | 1180 | 0.92 |
| 4 | GSTPCNGVEGFNCYFP | 476 | 0.91 |
| 4 | TVEKGIYQTSNFRVQP | 307 | 0.91 |
| 5 | GCLIGAEHVNNSYECD | 648 | 0.9 |
| 5 | LQSYGFQPTNGVGYQP | 492 | 0.9 |
| 5 | CCSCGSCCKFDEDDSE | 1247 | 0.9 |
| 6 | TEIYQAGSTPCNGVEG | 470 | 0.89 |
| 6 | TRFQTLLALHRSYLTP | 236 | 0.89 |
| 6 | YEQYIKWPWYIWLGFI | 1206 | 0.89 |
| 6 | HGVVFLHVTYVPAQEK | 1058 | 0.89 |
| 7 | IGKIQDSLSSTASALG | 931 | 0.88 |
| 7 | FAMQMAYRFNGIGVTQ | 898 | 0.88 |
| 7 | HVTYVPAQEKNFTTAP | 1064 | 0.88 |
| 8 | KQIYKTPPIKDFGGFN | 786 | 0.87 |
| 9 | CGPKKSTNLVKNKCVN | 525 | 0.86 |
| 9 | FERDISTEIYQAGSTP | 464 | 0.86 |
| 9 | YVGYLQPRTFLLKYNE | 266 | 0.86 |
| 9 | SWMESEFRVYSSANNC | 151 | 0.86 |
| 9 | CCSCLKGCCSCGSCCK | 1240 | 0.86 |
| 10 | TKTSVDCTMYICGDST | 732 | 0.85 |
| 10 | EVRQIAPGQTGKIADY | 406 | 0.85 |
| 10 | CFTNVYADSFVIRGDE | 391 | 0.85 |
| 10 | MSFPQSAPHGVVFLHV | 1050 | 0.85 |
Table 7: ABCpred prediction of B-cell epitopes.
Discussion
Phylogenetic analysis (Figure 1) shows that Indian strains along with the Asian strains for SARS-CoV-2 are distinct from the American and European strains, since they form separate clades as also stated by Forster. Although phylogenetically the virus forms distinct clades among its strains, the results of transition/ transversion ratio (Table 2) of spike glycoprotein implies that the virus is in a slow mutating state compare to other viruses like Zika or Influenza. Since spike protein acts as receptor binding and membrane fusion for entry inside the host cell, vaccines based on the spike protein could induce antibodies inside human host for inducing adaptive immunity. Among all the structural proteins of SARS-CoV-2, spike protein is the main antigenic component that will induce immune response inside the host and gives protective immunity against the viral infection by producing antibodies. So low mutation rate among the spike glycoproteins give hope for scientists to target it for corona vaccine design and antiviral development. If the virus evolves slowly, there will be a better chance for an effective vaccine for a long-term period against a wide range of population . Moreover, chances of the virus to form ‘escape mutants’ also diminishes due to having a smaller number of mutated versions of the SRAS-CoV-2 that are difficult to recognize by an induced vaccine. In compare to the Indian spike glycoprotein with rest of the countries, a clear evidence of having point mutation in the binding domain of the spike protein predicts more interaction with ACE2 receptor. But in case of United States, the spike glycoproteins show maximum variations between the sequences so far uploaded by the USA Govt. Furthermore from the amino acid composition point of view [Fig. III] we noticed more hydrophobic amino acids at the end region of the spike protein that may be the part of alphahelix spanning across the membrane . But a good proportion of hydrophilic regions on protein also predicts amino acids for solvent accessible and might be the good target regions for vaccine design. From T-cell and B-cell epitope prediction there are peptide segments within the Spike glycoprotein with good binding affinities for MHCs and able to generate antibodies within the human host for long term immunity. Currently, all countries are doing their hardest to tackle the infection against the pandemic and trying their best to come up with a potential vaccine against this deadly coronavirus, here our insilico study of the Indian spike glycoprotein provides insights and a list of mentioned epitope regions (Tables 5-7) that have a reasonable potential for vaccine design against SARS-CoV-2.
Conclusion
Since the spike surface glycoprotein is very important for viral entry and is part of the outer surface exposed structure of the viral capsid, our comparative study of the characteristics of the spike protein of the highly pathogenic human infecting coronavirus is very significant. From the graphical representation, hydropathy indices, transition/transversion ratios, amino acid composition, codon usage bias, protein properties we hypothesize that these could be the important parameters for protein morphology in further studies. Our study also gives new thinking for understanding the enhancement of viral pathogenicity and might explain in part the high incidence of infection cases being observed now. From the protein point of view, it is clear that there are regions that are surface exposed with solvent accessible and holds reasonable criteria to become potential epitopes for vaccines and can be augmented by appropriate adjuvants which lead to effective protection against SARS-CoV-2 infections.
Acknowledgement
We would like to thank Dr. Ashesh Nandy for his valuable corrections which helped to improve this paper.
Funding
The research work is not assisted by any kind of funding agencies or research fund.
Conflict of Interest
The authors have no conflict of interest.
References
- Roujian Lu, Xiang Zhao, Juan Li, Peihua Niu, Bo Yang, et al. (2020) Genomic characterization and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. The Lancet 395: 565-574.
- Venkatakrishnan K, Yalkinoglu O, Dong JQ (2020) Challenges in Drug Development Posed by the COVID‐19 Pandemic: An Opportunity for Clinical Pharmacology. Clinic. Pharmacol. Therapeutics.
- Peiris JSM, Lai ST, Poon LLM, Guan Yakan, Yam LYC, et al. (2003) Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361: 1319–1325
- Marra MA, Jones SJM, Astell CR (2000)The genome sequence of the SARS-associated coronavirus. Science 300:1399–1404.
- Rota PA, Oberste MS, Monroe SS (2003) Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394–1399.
- Zaki AM, Boheemen S, Bestebroer TM (2012) Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N. Engl. J. Med 367:1814–1820.
- Enjuanes L, Almazan F, Sola I (2006) Biochemical aspects of coronavirus replication and virus-host interaction. Annu. Rev. Microbiol 60: 211–230.
- Perlman S, Netland J (2019) Coronaviruses post-SARS: update on replication and pathogenesis. Nat. Rev. Microbiol 7: 439–50.
- Seah I, Agrawal R (2020) Can the Coronavirus Disease 2019 (COVID-19) Affect the Eyes? A Review of Coronaviruses and Ocular Implications in Humans and Animals. Ocul Immunol Inflamm 28: 391‐395.
- Tortorici MA, Veesler D (2019) Structural insights into coronavirus entry. Adv. Virus Res. 105: 93-116.
- De Haan CAM , Kuo L (1998) Coronavirus particle assembly: primary structure requirements of the membrane protein. J Virol 72: 6838-6850.
- Woo PCY, Huang Y, Lau, SKP (2010) Coronavirus genomics and bioinformatics analysis, Viruses. 2: 1804-1820.
- Yang D, Leibowitz JL (2015) The structure and functions of coronavirus genomic 30 and 50 ends. Virus. Res. 206:120.
- Lu, R., Zhao, X., Li, J., et al. 2020. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 395; 565-574.
- Hoffmann M, Kleine-Weber H, Schroeder S (2020) SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181: 271‐280.
- Guo Y-R, Cao Q-D., Hong, Z-S (2020) The origin, transmission and clinical therapies on coronavirus disease 2019 (COVID-19) out breakean update on the status. Mil Med Res 7:1-10.
- Nandy AA (1994) new graphical representation and analysis of DNA sequence structure: Methodology and Application to Globin Genes. Current Sci 66: 309-314.
- Raychaudhury C, Nandy A(1999) Indexing Scheme and Similarity Measures for Macromolecular Sequences. J. Chem.Infor.and Comp.Sci 39: 243-247.
- www.megasoftware.net
- http://emboss.open-bio.org/wiki/Appdocs
- Teresa Przytycka, PhD. Lecture 3 Predicting Transmembrene proteins and coiled coils Computational Aspects of Molecular
- Immune Epitode Database and Analysis Resource
- Saha S, Raghava GPS (2006) Prediction of Continuous B-cell Epitopes in an Antigen Using Recurrent Neural Network Proteins, PMID 65: 40-48.
- Duchen S, Holmes EC (2015) Declining transition/transversion ratios through time reveal limitations to the accuracy of nucleotide substitution models. BMC Evolutionary Biology 15:36.
- Dey S, Das S, Nandy A (2017) Characterization of Zika and Other Human Infecting Flavivirus Envelope Proteins and Determination of Common Conserved Epitope Region. EC Microbiology 8: 29-46.
- Im EH, Choi SS (2017) Synonymous Codon Usage Controls Various Molecular Aspects. Genomics & informatics. 15(4), 123–127.
- Purcell AW, McCluskey J, Rossjohn J (2007) More than one reason to rethink the use of peptides in vaccine design. Nat. Rev. 6: 404–414.
- Forstera P, Forsterd L, Renfrewb C (2020) Phylogenetic network analysis of SARS-CoV-2 genomes. PNAS 17: 9241–9243.
- Ferguson NM, Galvani AP Bush RM (2003) Ecological and immunological determinants of influenza evolution. Nature 422: 428-433.
- Hanley KA (2011) The double-edged sword: How evolution can make or break a live-attenuated virus vaccine. Evolution (NY) 4: 635‐643.
- Xu X, Chen P, Wang J, Feng J, Zhou H et al. (2020) Evolution of the novel coronavirus from the ongoing Wuhan outbreak and modeling of its Spike protein for risk of human transmission. Science China Life Sciences 63:457-60.
- Babcock GJ, Esshaki DJ, Thomas WD, Ambrosino DM( 2004) Amino acids 270 to 510 of the severe acute respiratory syndrome coronavirus spike protein are required for interaction with receptor. Journal of Virology 78:4552–4560.
- Robson B( 2020)COVID-19 Coronavirus spike protein analysis for synthetic vaccines, a peptidomimetic antagonist, and therapeutic drugs, and analysis of a proposed achilles’ heel conserved region to minimize probability of escape mutations and drug resistance. Comput Biol Med 11:103749
- Dey S, De A, Nandy A (2016) Rational Design of Peptide Vaccines against Multiple Types of Human Papillomavirus. Cancer Informatics15:1-16.
- Dey S, Nandy A, Basak SC (2017) A Bioinformatics approach to designing a Zika virus vaccine. Computational Biology and Chemistry 68:143-152.