Research Article, J Appl Bioinforma Comput Biol Vol: 7 Issue: 3
Bioinformatics Probing of Tissue-Specific MiRNAs Overexpressed in Active Tuberculosis Reveals Induction of Multiple Cancer Pathways
Elvis Ndukong Ndzi1-3, Ambily Nath IV4, Achuthsankar S Nair4*, Chellappan V Biju4, Shidhi PR4, Ousman Tamgue5, Alexis Ndjolo3, Jules Roger Kuiate1 and Celine Nguefeu Nkenfou3,6
1Department of Biochemistry, University of Dschang, Cameroon
2Laboratory of Animal Physiology and Health, Institute of Agricultural Research for Development (IRAD), Bambui, Cameroon
3Chantal Biya International Reference Center (CIRCB), Yaounde, Cameroon
4Department of Computational Biology and Bioinformatics, University of Kerala, Kerala, India
5University of Cape Town, Institute of Infectious Diseases and Molecular Medicine (IDM), Division of Immunology and South Africa Medical Research Council, Immunology of Infectious Disease, Faculty of Health Sciences Cape Town, South Africa
6Higher Teachers’ Training College, University of Yaounde I, Yaounde, Cameroon
*Corresponding Author : Achuthsankar S Nair
Department of Computational Biology and Bioinformatics, University of Kerala, Thiruvananthapuram-695581, Kerala, India
Tel: +91-4712308759
E-mail: sankar.achuth@gmail.com
Received: February 13, 2018 Accepted: June 18, 2018 Published: June 25, 2018
Citation: Ndzi EN, Nath IVA, Nair AS, Biju VC, Shidhi PR, et al. (2018) Bioinformatics Probing of Tissue-Specific MiRNAs Overexpressed in Active Tuberculosis Reveals Induction of Multiple Cancer Pathways. J Appl Bioinforma Comput Biol 7:3. doi: 10.4172/2329-9533.1000153
Abstract
This study attempts to discern the genes and pathways of tissue-specific miRNAs overexpressed in active tuberculosis (TB). The chosen 11 miRNAs were grouped into serum (S1), sputum (S2), and peripheral blood mononuclear cell (PBMC) (S3) classes. DIANA-miRPath v3 was used for the bioinformatics analyses. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) practices were used to pinpoint the key regulatory channels and functional annotations associated with the latent state. ECMreceptor interaction, transcriptional misregulation in cancer, nonsmall cell lung cancer, proteoglycans, and pathways in cancer were identified as the prime pathways in TB-related miRNAs. S1 and S3 transcripts showed the unifying ECM-receptor interaction with collagen (COL) and laminin (LAM) genes common to both datasets. Further research on these cancer-related cascades could provide mechanistic and therapeutic insights into TB pathophysiology.
Keywords: MiRNAs; Overexpressed; Active tuberculosis; Bioinformatics
Introduction
According to World Health Organization (WHO), one third of world’s population is infected with tuberculosis (TB) and about 10% of those infected (latent TB) progress to active TB though this percentage increases with the burden of immunocompromised persons [1]. TB, caused by Mycobacterium tuberculosis (Mtb) is an airborne disease commonly affects the lungs (pulmonary TB) but can also attack other body tissues (extrapulmonary TB). TB infection begins with the inhalation of aerosol droplets containing mycobacteria. They reach alveolar space and phagocytosed by macrophages which in turn induce the innate immune response. The macrophages, along with Mtb, and host defense cells form organized structures called granuloma, the hallmark of TB. They eventually rupture releasing the infectious bacilli to airways or other tissues [2,3].
MicroRNAs (miRNAs) are short (~22 nucleotides) non-coding RNAs that regulate gene expression post-transcriptionally. They bind to target messenger RNAs (mRNAs) and repress protein production by mRNA degradation or temporal silencing. These binding usually take place between positions 2-8 (“seed”) of miRNA and 3’ untranslated region (UTR) of mRNA though some binding seldom occur at coding sequence (CDS) and 5’UTR of mRNA [4]. MiRNAs are implicated in a wide range of disease including diabetes, cancer, TB, cardiovascular, and neurological diseases. Their altered expression discriminate the pathologic states of a diseased person from that of a healthy control. Hence, they are used as excellent biomarkers [5].
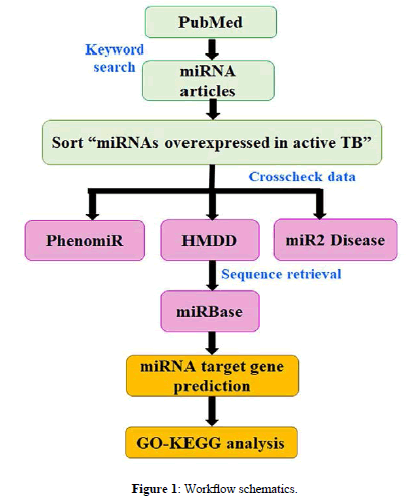
The present study aims to provide hypothetical insights into the signaling pathways induced by serum, sputum, and PBMC miRNAs overexpressed in active TB. We applied a bioinformatics pipeline to delineate the similarity/dissimilarity in the genes and pathway patterns of upregulated miRNAs of the above three tissues. A schematic representation of present work is described in Figure 1.
Materials and Methods
Data source
A systematic PubMed search done with the queries ‘miRNAs associated with tuberculosis, miRNAs related to tuberculosis, miRNAs+tuberculosis, and miRNAs as potential biomarkers for tuberculosis’, indexed 282 records. This data sorted to retain 11 upregulated miRNAs in active TB and are compiled in 1) PhenomiR 2.0 (http://mips.helmholtz-muenchen.de/phenomir) [6], 2) HMDD 2.0 (http://www.cuilab.cn/hmdd) [7], and 3) miR2Disease (http://www.miR2Disease.org) [8]. The sequences were accessed from miRBase Release 21 (www.mirbase.org) [9] and ensured their accuracy by miRBase Tracker (www.mirbasetracker.org) [10].
GO-KEGG annotation and target gene prediction
All computational calculations were done with DIANA-miRPath v3.0 (http://www.microrna.gr/miRPathv3) [11]. The KEGG/GO analysis module was chosen for the respective functional analysis with microT-CDS v5, TarBase v7, and TargetScan v6 used for target gene prediction by keeping their default cut-off parameters. The results were decoded by ‘Pathways/Categories Union’ option of Posteriori Analysis Method. MicroT-CDS and TargetScan results correspond to predicted miRNA-mRNA interactions while TarBase incorporated experimentally validated information.
Results
Grouping of tissue-specific TB miRNAs
Our literature review (2010-2017) provided 31 TB-related miRNAs, of which, 11 members found to be upregulated in active TB (versus healthy controls) formed the dataset for present study (Table 1). They were grouped by their tissue-specific induction into 1) serum (S1), 2) sputum (S2), and 3) PBMC (peripheral blood mononuclear cells) (S3) transcripts. S1 composed of hsa-miR-29a-3p, hsa-miR- 361-5p, hsa-miR-889-5p, hsa-miR-576-3p, hsa-miR-196b-5p, and hsa-miR-376c-3p. S2 group included hsa-miR-3179 and hsa-miR- 147a while S3 members were hsa-miR-29a-3p, hsa-miR-144-5p, hsamiR- 155-5p, and hsa-miR-155-3p.
| miRNA | miRBase accession |
Mature sequence | Reference | Length (nt) |
Mol.wt (kDa) |
GC (%) | ΔG (Kcal/mol) |
|---|---|---|---|---|---|---|---|
| hsa-miR-29a-3p | MIMAT0000086 | UAGCACCAUCUGAAAUCGGUUA | [12-15] | 22 | 6634.3 | 41 | 27.1 |
| hsa-miR-361-5p | MIMAT0000703 | UUAUCAGAAUCUCCAGGGGUAC | [16,13] | “ | 6650.3 | 45 | 26.3 |
| hsa-miR-144-5p | MIMAT0004600 | GGAUAUCAUCAUAUACUGUAAG | [17] | “ | 6659.3 | 32 | 22 |
| hsa-miR-155-5p | MIMAT0000646 | UUAAUGCUAAUCGUGAUAGGGGU | [18] | 23 | 7021.5 | 39 | 27.8 |
| hsa-miR-155-3p | MIMAT0004658 | CUCCUACAUAUUAGCAUUAACA | [18] | 22 | 6539.2 | 32 | 23.4 |
| hsa-miR-3179 | MIMAT0015056 | AGAAGGGGUGAAAUUUAAACGU | [19] | “ | 6801.4 | 36 | 26.5 |
| hsa-miR-147a | MIMAT0000251 | GUGUGUGGAAAUGCUUCUGC | [19] | 20 | 6080.9 | 50 | 25.2 |
| hsa-miR-889-5p | MIMAT0026719 | AAUGGCUGUCCGUAGUAUGGUC | [16] | 22 | 6683.3 | 50 | 28.3 |
| hsa-miR-576-3p | MIMAT0004796 | AAGAUGUGGAAAAAUUGGAAUC | [16] | “ | 6785.4 | 32 | 24.4 |
| hsa-miR-196b-5p | MIMAT0001080 | UAGGUAGUUUCCUGUUGUUGGG | [20] | “ | 6678.2 | 45 | 27.1 |
| hsa-miR-376c-3p | MIMAT0000720 | AACAUAGAGGAAAUUCCACGU | [20] | 21 | 6391.2 | 38 | 24.9 |
Table 1: Sequence details and 2D descriptors of 11 miRNAs overexpressed in active tuberculosis.
MiRNA-mRNA network prediction
Target genes of each miRNA class were individually defined by microT-CDS, TarBase, and TargetScan. MicroT-CDS and TargetScan provided predicted miRNA-gene interactions while TarBase documented experimentally validated records. For S1, 2400, 3168, and 3259 genes were identified by above three tools respectively (Supplementary Tables S1, S2, S3). The S2 gene statistics were as follows: microT-CDS- 1044; TarBase- 467; TargetScan-314 (Supplementary Tables S4-S6). S3-specific 1424 (microT-CDS), 2598 (TarBase), and 403 (TargetScan) genes were recognised (Supplementary Tables S7-S9).
Putative target genes for S1-S2-S3 miRNAs were enlisted by microT-CDS and TargetScan. For S1 hsa-miR-29a-3p, hsa-miR- 361-5p, hsa-miR-889-5p, hsa-miR-576-3p, hsa-miR-196b-5p, and hsa-miR-376c-3p, 339, 465, 380, 214, 207, 308 (microT-CDS) (Supplementary Table S10), 229, 19, 99, 28, 64, and 48 genes were identified respectively (TargetScan) (Table S3). The S2 hsa-miR-3179 and hsa-miR-147a gene counts were as follows: 331, 673 (microTCDS) (Supplementary Table S11), 238, and 76 genes respectively (TargetScan) (Table S6). S3-specfiic hsa-miR-29a-3p, hsa-miR-144- 5p, hsa-miR-155-5p, and hsa-miR-155-3p inferred to bind to 339, 92, 373, 137 (microT-CDS) (Supplementary Table S12), 229, 30, 49, and 95 genes respectively (TargetScan) (Table S9).
GO annotation
The GO annotation such as biological process (BP), molecular function (MF), and cellular component (CC) of S1-S2-S3 completed with the first three functional terms selected for scrutinization. From these, the top most GO terms enriched by each miRNA class is discussed.
The microT-CDS-predicted S1 targets presented cellular nitrogen compound metabolic process (GO:0034641), ion binding (GO:0043167), and organelle (GO:0043226) (Supplementary Table S13). TarBase-based S1 genes took part in nucleobase-containing compound catabolic process (GO:0034655), protein binding transcription factor activity (GO:0000988), and cellular_component (GO:0005575) (Supplementary Table S14). Extracellular matrix disassembly (GO:0022617), extracellular matrix structural constituent (GO:0005201), and endoplasmic reticulum lumen (GO:0005788) (Supplementary Table S15) were the GO terms determined by TargetScan.
As shown in Supplementary Table S16, S2 genes promoted synaptic transmission (GO:0007268), ion binding (GO:0043167), and organellar-based (GO:0043226) functionalities (microT-CDS). TarBase indicated that these sputum miRNAs are involved in biosynthetic process (GO:0009058), ion binding (GO:0043167), and organellar-located (GO:0043226) (Supplementary Table S17). The TargetScan-identified genes presumed to perform homophilic cell adhesion via plasma membrane adhesion molecules (GO:0007156) along with ion binding (GO:0043167) and are organellar-originated (GO:0043226) (Supplementary Table S18). The microT-CDS gene annotation of S3 (Supplementary Table S19) was similar to S1, i.e., BP: cellular nitrogen compound metabolic process, MF: ion binding, CC: organelle. As described in (Supplementary Table S20), TarBase grouped the PBMC-specific TB transcripts to be involved in blood coagulation (GO:0007596), RNA binding (GO:0003723), and are cellular component-based (GO: 0005575). TargetScan predicted extracellular matrix disassembly (GO: 0022617), ion binding, and endoplasmic reticulum lumen (GO: 0005788) as the top most terms (Supplementary Table S21).
KEGG pathway enrichment
The KEGG analysis of S1 target genes found COL and LAM genes significantly enriched the top most ECM-receptor interaction (hsa04512) (Table 2) (Supplementary Table S22). The microT-CDSderived S2 genes involved in drug addiction (morphine (hsa05032), nicotine (hsa05033)) and biosynthesis of unsaturated fatty acids (hsa01040) (Table 3); GABA gene participation in these pathways is noted (Supplementary Table S23). The S2 genes (characterized by TarBase) enriched transcriptional misregulation in cancer (hsa05202), protein processing in endoplasmic reticulum (hsa04141), and non-small cell lung cancer (hsa05223); TargetScan-predicted S2 genes showed to regulate lipid and purine metabolic pathways. The S3 genes (Table 4) formulated by microT-CDS, TarBase, and TagetScan showed the unifying ECM-receptor interaction (hsa04512) where COL and LAM form the major gene constituent (Supplementary Table S24). We hereby conclude with TarBase GO/KEGG results as its predictions are based on experimentally proven miRNA-mRNA interactions.
| MicroT-CDS prediction | ||||
|---|---|---|---|---|
| No | KEGG pathway | MiRNA | P-value | Gene count |
| 1 | ECM-receptor interaction (hsa04512) | hsa-miR-29a-3p hsa-miR-196b-5p has-miR-376c-3p |
8.967E-116 4.25E-07 0.002789058 |
18 3 4 |
| 2 | Amoebiasis (hsa05146) | hsa-miR-29a-3p | 2.77E-16 | 20 |
| 3 | Protein digestion and absorption (hsa04974) | hsa-miR-29a-3p | 3.61E-10 | 23 |
| TarBase prediction | ||||
| 1 | ECM-receptor interaction (hsa04512) | hsa-miR-29a-3p hsa-miR-361-5p has-miR-196b-5p |
1.54E-53 0.026815661 0.001646394 |
22 5 8 |
| 2 | Fatty acid biosynthesis (hsa00061) | hsa-miR-29a-3p | 2.84E-18 | 2 |
| 3 | Lysine degradation (hsa00310) | hsa-miR-29a-3p hsa-miR-361-3p hsa-miR-576-3p hsa-miR-196b-5p |
7.28E-05 0.004402843 2.25E-08 0.01174639 |
10 5 7 6 |
| TargetScan prediction | ||||
| 1 | ECM-receptor interaction (hsa04512) | hsa-miR-29a-3p | 7.43E-96 | 12 |
| 2 | Amoebiasis (hsa05146) | hsa-miR-29a-3p | 5.71E-14 | 13 |
| 3 | Morphine addiction (hsa05032) | hsa-miR-361-5p hsa-miR-376c-3p |
1.43E-11 0.023530296 |
2 1 |
Table 2: Top 3 KEGG pathways enriched by target genes of at least one miRNA of S1 class predicted by microT-CDS, TarBase, and TargetScan.
| MicroT-CDS prediction | ||||
|---|---|---|---|---|
| No | KEGG pathway | MiRNA | P-value | Gene count |
| 1 | Morphine addiction (hsa05032) | hsa-miR-147a | 1.46E-08 | 10 |
| 2 | Nicotine addiction (hsa05033) | hsa-miR-147a | 4.49E-06 | 7 |
| 3 | Biosynthesis of unsaturated fatty acids (hsa01040) | hsa-miR-3179 | 2.66E-05 | 1 |
| TarBase prediction | ||||
| 1 | Transcriptional misregulation in cancer (hsa05202) | hsa-miR-3179 | 1.94E-05 | 8 |
| 2 | Protein processing in endoplasmic reticulum (hsa04141) | hsa-miR-317 | 0.00459851 | 12 |
| 3 | Non-small cell lung cancer (hsa05223) | hsa-miR-3179 | 0.00459851 | 6 |
| TargetScan prediction | ||||
| 1 | Biosynthesis of unsaturated fatty acids (hsa01040) | hsa-miR-3179 | 6.12E-08 | 1 |
| 2 | Glycerophospholipid metabolism (hsa00564) | hsa-miR-147a | 0.000294033 | 2 |
| 3 | mRNA surveillance pathway (hsa03015) | hsa-miR-3179 | 0.005126026 | 5 |
Table 3: Top 3 KEGG pathways enriched by target genes of at least one miRNA of S2 class predicted by microT-CDS, TarBase, and TargetScan.
| MicroT-CDS prediction | ||||
|---|---|---|---|---|
| No | KEGG pathway | MiRNA | P-value | Gene count |
| 1 | ECM-receptor interaction (hsa04512) | hsa-miR-29a-3p | 8.97E-116 | 18 |
| 2 | Amoebiasis (hsa05146) | hsa-miR-29a-3p | 2.77E-16 | 20 |
| 3 | Protein digestion and absorption (hsa04974) | hsa-miR-29a-3p | 3.61E-10 | 23 |
| TarBase prediction | ||||
| 1 | ECM-receptor interaction (hsa04512) | hsa-miR-29a-3p | 1.54E-53 | 22 |
| 2 | Fatty acid biosynthesis (hsa00061) | hsa-miR-29a-3p | 2.84E-18 | 2 |
| 3 | Viral carcinogenesis (hsa05203) | hsa-miR-29a-3p | 7.46E-10 | 42 |
| TargetScan prediction | ||||
| 1 | ECM-receptor interaction (hsa04512) | hsa-miR-29a-3p | 7.43E-96 | 12 |
| 2 | Amoebiasis (hsa05146) | hsa-miR-29a-3p | 5.71E-14 | 13 |
| 3 | Protein digestion and absorption (hsa04974) | hsa-miR-29a-3p | 8.86E-10 | 16 |
Table 4: Top 3 KEGG pathways enriched by target genes of at least one miRNA of S3 class predicted by microT-CDS, TarBase, and TargetScan.
Discussion
Characterization of the functional pathways of tissue-specific miRNAs overexpressed in latent infection could confer novel insights into pathophysiology and therapy of TB. Our study ascertained the distinct gene-pathway regulatory network of serum (S1), sputum (S2), and PBMC (S3) miRNAs upregulated in active TB. We observed that the top ranked GO BP and MF of each miRNA class found to differ each other. In S1, the nucleobase-containing compound catabolic process was induced by hsa-miR-29a-3p, hsa-miR-361-5p, and hsa-miR-196b-5p that share G3BP, AGO, HSP, DDX, RAB, and CNOT genes in common (Supplementary Table S14). The biosynthetic process of S2 was modulated by 132 unique genes of hsa-miR-3179. Blood coagulation induced by hsa-miR-29a-3p and hsa-miR-155-5p in S3 showed overlapping of CALU, SPARC, GLG, THBS, and ATP genes (Supplementary Table S20).
The KEGG analysis emphasized the intriguing link between TB miRNAs and cancer cascades. ECM-receptor interaction was defined as the prime S1-S3 pathway where the ECM molecules such as COL and LAM form the major enriched genes (Supplementary Tables S22 and S24). This cancer pathway found to have a key role in TB as its disruption causes matrix degradation and imbalanced immune response that lead to pulmonary cavitation, the sign of TB infection [21]. S2 genes found to promote transcriptional misregulation in cancer, non-small cell lung cancer, proteoglycans, and pathways in cancer. All these predictions were accurate as they agreed well with previous findings that ECM-receptor interaction [14,21] and lysine degradation [22] have crucial roles in promoting TB pathogenesis. A recent publication disclosed the coexistence of TB and lung cancer [23].
The analogous KEGG functions of S1 and S3 accounted for hsamiR- 29a-3p, common to both miRNA classes. Also, these transcripts are derivatives of blood tissue components (S1 of serum and S3 of PBMC). Hsa-miR-29a-3p genes enriched all the predicted S1-S3 pathways such as ECM-receptor interaction, fatty acid catabolism, viral carcinogenesis, and adherens junction. These results supplement the previous findings [12,13] that miR-29a-3p is a stable potential biomarker of active TB.
Conclusion
Bioinformatics-based functional annotation of tissue-specific miRNAs overexpressed in active TB could provide valuable clues into the pathogenic mechanism. The induction of ECM-receptor interaction, transcriptional misregulation in cancer, non-small cell lung cancer, proteoglycans, and pathways in cancer by the latent TB effectors were very interesting. An in-depth experimental study is recommended to unravel the role of these cancer cascades in TBspecific miRNAs. The S1-S3 analogy at pathway (ECM-receptor interaction) and gene (COL and LAM as dominant group) levels could be attributed to their blood tissue-origin and sharing of hsa-miR- 29a-3p in common. The identified putative genes could shed light on the structural, functional, and therapeutic aspects of overexpressed miRNAs in active TB.
Acknowledgements
This project was funded by C.V. Raman International Fellowship of African Researchers under Visiting Fellowship (Research Grant: INT/NAI/CVRF/2014). We thank the Department of Science and Technology (DST), India, Federation of Indian Chambers of Commerce and Industry (FICCI) and Government of India for granting this prestigious award. We are indebted to Department of Computational Biology & Bioinformatics, Thiruvananthapuram, for providing the infrastructure for conducting the work.
Conflicts of Interest
The authors declare no conflicts of interest regarding the publication of this work.
Author Contributions
Conception and design, analysis and interpretation of data, manuscript preparation: ENN, ANIV. Supervision of the project and manuscript correction: ASN. Manuscript correction and creative suggestions: BC, SPR. Manuscript preparation: OT, AN, KJR. Study conception, design and manuscript preparation: CNN.
References
- World Health Organization (2017) Tuberculosis Fact sheet Reviewed March.
- Kumar V, Abbas AK, Fausto N, Mitchell RN (2007) Robbins Basic Pathology. (8th edtn.), Saunders Elsevier 516-522.
- Khan (2011) Essence Of Paediatrics. Elsevier India, 401.
- Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136: 215-233.
- van Rooij E, Kauppinen S (2014) Development of microRNA therapeutics is coming of age. EMBO Mol Med 6: 851-864.
- Ruepp A, Kowarsch A, Schmidl D, Buggenthin F, Brauner B, et al. (2010) PhenomiR: a knowledge base for miRNA expression in diseases and biological processes. Genome Biol 11: R6.
- Lu M, Zhang Q, Deng M, Miao J, Guo Y, et al. (2008) An analysis of human microRNA and disease associations. PLoS One 3: e3420.
- Jiang Q, Wang Y, Hao Y, Juan L, Teng M, et al. (2009) miR2Disease: a manually curated database for microRNA deregulation in human disease. Nucleic Acid Res 37: D98-D104.
- Jones GS, Grocock RJ, van Dongen S, Bateman A, Enright AJ (2006) miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res 34: D14-D144.
- Van Peer G, Lefever S, Anckaert J, Beckers A, Rihani A, et al. (2014) miRBase Tracker: keeping track of microRNA annotation changes. Database (Oxford) pii: bau080
- Vlachos IS, Zagganas K, Paraskevopoulou MD, Georgakilas G, Karagkouni D (2015) DIANA-miRPath v3.0: deciphering microRNA function with experimental support. Nucleic Acids Res 43: W460-W466.
- Fu Y, Yi Z, Wu X, Li J, Xu F (2011) Circulating microRNAs in patients with active pulmonary tuberculosis. J Clin Microbiol 49: 4246-4251.
- Nehal ID, Soha AR, Marwa SE, Emad EA, Nagwa MA (2014) Serum microRNA-29a and microRNA-361-5p as potential diagnostic biomarkers for active pulmonary tuberculosis. Egypt J Med Microbiol 23: 27-35.
- Fu Y, Yi Z, Li J, Li R (2014) Deregulated microRNAs in CD4+T cells from individuals with latent tuberculosis versus active tuberculosis. J Cell Mol Med 18: 503-513.
- Zhou M, Yu G, Yang X, Zhu C, Zhang Z, et al. (2016) Circulating microRNAs as biomarkers for the early diagnosis of childhood tuberculosis infection. Mol Med Rep 13: 4620-4626.
- Qi Y, Lunbiao C, Yiyue G, Zhiyang S, Kangchen Z, et al. (2012) Altered serum microRNAs as biomarkers for the early diagnosis of pulmonary tuberculosis infection. BMC Infect Dis 12: 384.
- Liu Y, Wang X, Jiang J, Cao Z, Yang B, Cheng X (2011) Modulation of T cell cytokine production by miR-144 * with elevated expression in patients with pulmonary tuberculosis. Mol Immunol 48: 1084-1090.
- Wu J, Lu C, Diao N, Zhang S, Wang S, et al. (2012) Analysis of microRNA expression profiling identifies miR-155 and miR-155 * as potential diagnostic markers for active tuberculosis: a preliminary study. Human Immunol 73: 31-37.
- Yi Z, Fu Y, Ji R, Li R, Guan Z (2012) Altered microRNA signatures in sputum of patients with active pulmonary tuberculosis. PLoS One 7: e43184.
- Zhang H, Sun Z, Wei W, Liu Z, Fleming J, et al. (2014) Identification of serum microRNA biomarkers for tuberculosis using RNA-seq. PLoS One 9: e88909.
- Brilha S, Wysoczanski R, Whittington AM, Friedland JS, Porter JC (2017) Monocyte adhesion, migration, and extracellular matrix breakdown are regulated by integrin αVb3 in Mycobacterium tuberculosis infection. J Immunol 199: 982-991.
- Alipoor SD, Mortaz E, Tabarsi P, Farnia P, Mirsaeidi M, et al. (2017) Bovis Bacillus Calmette-Guerin (BCG) infection induces exosomal miRNA release by human macrophages. J Transl Med 15: 105.
- Barh D, Tiwari S, Kumavath RN, Azevedo V (2017) A potential link between tuberculosis and lung cancer through non-coding RNAs. Biorxiv.