Case Report, Genet Disor Genet Rep Vol: 11 Issue: 6
Clinical and Genetic Study of 1p36 Deletion Syndrome in Three Tunisian Patients
Yasmina Elaribi*, Houweyda Jilani, Syrine Hizem, Imen Rejeb, Lilia Kraoua, Caroline Rooryck-Thambo, Ahmed Maherzi, Ridha Mrad and Lamia Ben Jemaa
Department of Genetic Science, Mongi Slim Hospital, Tunis, Tunisia
*Corresponding Author: Yasmina Elaribi Department of Genetic Science, Mongi Slim Hospital, Tunis, Tunisia, E-mail: yasmina.elaribiatrns.tn
Received: 07 October, 2019, Manuscript No. JGDGR-19-3317; Editor assigned: 10 October, 2019, PreQC No. JGDGR-19-3317 (PQ); Reviewed: 24 October, 2019, QC No. JGDGR-19-3317; Revised: 29 July, 2022, QI No. JGDGR-19-3317; Manuscript No. JGDGR-19-3317 (R); Published: 26 August, 2022, DOI:10.4172/2327-5790.1000031
Citation: Elaribi Y, Jilani H, Hizem S, Rejeb I, Kraoua L, et al . (2022) Clinical and Genetic Study of 1p36 Deletion Syndrome in Three Tunisian Patients. Genet Disor Genet Rep 11:6.
Abstract
Purpose: 1p36 deletion syndrome is a clinically recognizable syndrome characterized by typical dysmorphic features such as straight eyebrows, deep-set eyes, midface hypoplasia, orofacial clefting, pointed chin and low-set ears. It is also associated with developmental delay, microcephaly, hypotonia and growth retardation. Cardiac malformations, hearing loss and ophthalmologic anomalies are also described. To date, many cases have been reported worldwide. However, this syndrome had never been reported in Tunisian population. We describe in this study the clinical and molecular characteristics of three Tunisian patients with the 1p36 deletion syndrome.
Patients and methods: The clinical characteristics of the three patients were reviewed. Karoytypes, array CGH and FISH were performed for genetic diagnosis.
Results: All patients had brachymicrocephaly, development delay and growth retardation. Two of them had typical dysmorphic features consisting of straight eyebrows, midface hypoplasia, pointed chin and low-set ears. The other patient had orofacial clefting. Karyotypes were normal in all cases. Array CGH showed 1p36 monosomy in all patients. Besides, it revealed 22q13 trisomy in one patient whose father had a translocation t (1;22). These findings were validated by FISH.
Conclusion: The 1p36 deletion syndrome is a common malformation syndrome that is not always clinically recognizable. Array CGH was a significant tool for patients in whom the diagnosis was not suspected.
Keywords: 1p36 Deletion syndrome; Monosomy 1p36; Array comparative genomic hybridization; Fish; Paternal translocation
Introduction
1p36 deletion syndrome, also called monosomy 1p36 is a common microdeletion syndrome with an estimated incidence of 1 in 5000 to 1 in 10000. It is characterized by typical dysmorphic features such as straight eyebrows, deep-set eyes, down-slanting palpebral fissures, midface hypoplasia, flat nasal bridge, orofacial clefting, pointed chin and posteriorly rotated, low-set ears. It is also associated with developmental delay, microbrachycephaly, hypotonia, seizures and growth retardation. Cardiac malformations are seen in 71% of cases including 23% of noncompaction cardiomyopathy. Sensorineural hearing loss and ophthalmologic anomalies are seen in 28% and 52% of patients respectively. Brain malformation, feeding difficulties and hypothyroidism are also described. 1p36 deletion syndrome has never been reported in the Tunisian population. In this study, we describe the clinical features and molecular findings in three Tunisian patients with this syndrome [1].
Case Series
Patients and methods
Three female patients were diagnosed with 1p36 deletion syndrome in the genetic department of Mongi Slim Hospital, Tunis, between 2012 and 2017. They had physical examination and radiological investigations.
Informed consent was obtained from the patients’ parents. We applied conventional cytogenetic analysis (R- and or G-banding) of peripheral blood lymphocytes at approximately 400-550 band resolution per haploid set. DNA was extracted from peripheral blood using standard methods. Oligonucleotide-based 8 × 60 K array-Comparative Genomic Hybridization (CGH) using agilent platform was performed following standard and manufacturer’s recommendations. Confirmation was made by Fluorescence In situ Hybridization (FISH) analysis [2].
Results
Case 1
The patient was the second child of healthy, non-consanguineous parents. She had a normal six-year-old sister. There was no familial history of malformation or intellectual disability. Obstetric ultrasounds showed hydrocephalus that was not explored by other investigations. She was born at term by cesarean section with a birth weight of 2600 g (3rd percentile), length of 48 cm (50th percentile) and Occipital-Frontal Circumference (OFC) of 32 cm (3rd percentile). She was admitted at the neonatology department for cyanosis and bilateral orofacial cleft. Patent Ductus Arteriosus (PDA) and Ventricular Septal Defect (VSD) were found in cardiac ultrasound, for which she was undergoing medical treatment. Abdominal ultrasound was normal [3].
At the age of four months, she was admitted at pediatric department for failure to thrive, hypotrophy (weight at 3800 g, <3rd percentile) and hypotony. She had dysmorphic features including hypertelorism, epicanthal folds, left ptosis, long eyelashes, depressed nasal bridge, bilateral oro-facial cleft and rethrognatia. She also had umbilical hernia, coccyx hair tuft and overlapping toes. She was treated by Phenobarbital for epilepsy. At the age of one year, she was diagnosed with hypothyroidism. Because of speech delay, auditory exploration was performed and revealed hearing loss. Cerebral MRI didn’t reveal any malformation but a moderate cortical atrophy. Her psychomotor development was retarded; she could not sit at the age of four.
Karyotype was normal. Because of the association of clefting and congenital cardiopathy, FISH analysis for DiGeorge syndrome at 22q11 and 10p14 was made but showed no anomaly. Array CGH revealed a deletion of 8.7 Mb in the 1p36 region, containing 146 genes. This investigation was confirmed by FISH analysis [4].
Case 2
The patient was the second child of healthy, non-consanguineous parents. She had a three-year-old brother with normal psychomotor development. There was a history of intellectual disability in two paternal aunts. She was born at term by vaginal delivery after an uneventful pregnancy. She had a birth weight of 2725 g (15th percentile), length of 48 cm (50th percentile) and OFC of 33 cm (15th percentile). She was referred to the genetic department at the age of 11 months for psychomotor delay and failure to thrive. She had gastro-esophageal reflux due to cardia malposition [5].
The examination revealed dysmorphic features with narrow and hirsute forehead, synophris, downslanting eyebrows and palpebral fissures, deep-set eyes, long eyelashes, anteverted nares, microstomia, low-set ears and short neck. She had bilateral overlapping toes, camptodactyly of big toes, joint laxity and hypertrichosis of the back and thighs [6].
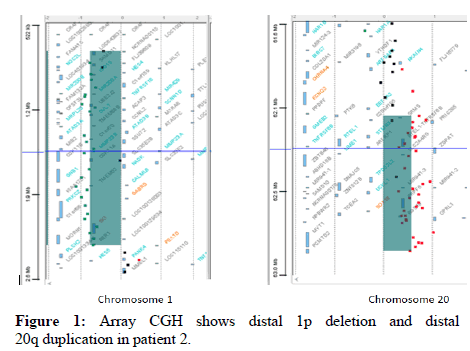
Cerebral MRI, cardiac ultrasounds and karyotype were normal. CGH revealed a deletion of 1.57 Mb in the 1p36 region, besides of a duplication of 730 kb in 20q13. This anomaly was due to a paternal translocation between chromosomes 1 and 20 revealed by FISH (Figure 1).
Case 3
The patient was the second child of healthy, non-consanguineous parents. She had an eight-year-old sister in a good health. Pregnancy was complicated by toxoplasmosis seroconversion which was treated for two months. She was born at term by cesarean section for fetal distress. She had a birth weight of 2560 g (3rd percentile). Length and OFC were not mentioned. She was admitted at the neonatology department for neonatal respiratory distress. Cardiac ultrasounds revealed left-to-right shunt and pulmonary hypertension which was operated at the age of two years and a half. She had clonic seizures at the age of two months, with good evolution under Diazepam [7-9].
She was referred to genetic department at the age of 4 years and a half for suspicion of Smith-Magenis syndrome because of self-injury, low sensitivity to pain, sleep disturbances and disorder of circadian timing. The physical examination showed dysmorphic features: Elongated face, downslanting, straight eyebrows, strabismus, short philtrum, thin lips, pointed chin, small hands, overlappings toes with camptodactyly of big toes. She had central hypotonia and severe development delay: She could not stand alone and did not say any word [10-14].
Karyotype was normal. Array CGH revealed a deletion of 7.27 Mb in the 1p36 region. This investigation was confirmed by FISH analysis (Table 1).
| Variable | Patient n°1 | Patient n°2 | Patient n°3 |
|---|---|---|---|
| Age at first consultation | 4 months | 11 months | 4 years ½ |
| Birth | |||
| Gestational age | At term | At term | At term |
| Weight (percentile) | 2600 (15th) | 2725 (15th) | 2560 (3rd) |
| Height (percentile) | 48 (15th) | 48 (15th) | NA |
| OFC (percentile) | 32 (3rd) | 33 (15th) | NA |
| APGAR score 1 min/5 min | 08-Sep | 10-Oct | 06-Aug |
| Typical dysmorphic features | - | + | + |
| Microbrachycephaly | - | + | + |
| Straight eyebrows | - | + | + |
| Deep-set eyes | - | + | - |
| Downslanting palpebral fissures | - | + | - |
| Flat nasal bridge | + | + | + |
| Midface hypoplasia | + | + | - |
| Pointed chin | - | + | + |
| Posteriorly rotated, low-set ears | - | + | - |
| Orofacial cleft | + | - | - |
| Developmental findings | |||
| Developmental delay | + | + | + |
| Mental retardation | + | + | + |
| Poor language | + | + | + |
| Walk | - (5 years) | - (3 years) | - (5 years) |
| Behavioral anomalies | Autoaggression | - | Autoaggression Sleep disturbance |
| Neurologic findings | |||
| Hypotonia | + | + | + |
| Seizures | + | + | + |
| Cerebral MRI | Moderate cortical atrophy | Normal | Normal |
| Auditory exploration | Bilateral deafness | NA | Bilateral deafness |
| Visceral anomalies | |||
| Heart | PDA, VSD | Normal | Left-to-right shunt , PH |
| Gastro-intestinal tract | NA | GER | Normal |
| Kidneys | Normal | Normal | Normal |
| Limbs/Skeleton defects | Overlapping toes | Overlapping toes camptodactyly | Overlapping toes camptodactyly scoliosis |
| Skin | Hypertrichosis | Hypertrichosis | Normal |
| Hypothyroidism | + | NA | NA |
| Karyotype | 46,XX | 46,XX | 46,XX |
| CGH array | 8.7 Mb deletion in 1p36.33p36.23 (759,762-9,503,580 hg19) | 1.57 Mb deletion in 1p36.33p36.32 (759,762-2,335,155 hg19) 730 kb duplication in 20q13.33 (62,163,415-62,893,189 hg19) | 6.5 Mb deletion in 1p36.33p36.23 (759,762-7,279,185 hg19) |
| Parents FISH | NA | Paternal t (1;20) | Normal |
Table 1: Clinical and genetic findings of the three patients. Note: NA: Not Available, OFC: Occipito-Frontal Circumference, PDA: Patent Ductus Arteriosus, PH: Pulmonary Hypertension, VSD: Ventricular Septal Defect
Discussion
1p36 deletion syndrome is considered as the most common terminal deletion syndrome. In this study, we describe the clinical and molecular characteristics of three Tunisian patients diagnosed with 1p36 monosomy [15].
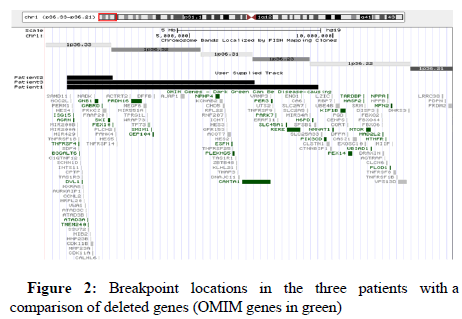
It has been reported that all patients have distinctive dysmorphic features, intellectual disability and diffuse hypotonia. Because patient P1 showed bilateral orofacial clefting and congenital cardiopathy, she was clinically misdiagnosed as having DiGeorge syndrome. A critical region for cleft palate in 1p36 deletion syndrome was defined by Shimada who found cleft palate and/or cleft lip in 14/50 patients. This region contains the v-ski sarcoma viral oncogene homolog (SKI) gene which haploinsufficiency is thought to contribute, besides to clefting, to the developmental delay, intellectual disability, seizures and congenital heart defects. All subjects that show cleft lip and/or cleft palate are deleted for the SKI gene. As expected, this gene was deleted in our three patients. Because it is located in the most distal 2Mb of 1p, most subjects are deleted for this gene, but only 17% show clefting abnormalities [16] (Figure 2).
As in previously reported cases, developmental delay and intellectual deficiency were noticed in all patients of this study although no psychological test was used. Severe to profound mental retardation was reported in 88% of patients. Expressive language was absent in 75% of cases, limited to one or two words in the rest of the cases [17].
As it was noted in our study, there is no correlation between the deletion size and the number of observed clinical features: even subjects with relatively small deletions (<3 Mb) have most of the features associated with monosomy 1p36. Thus, most genes associated with the phenotype of monosomy 1p36 would be at the distal end of the chromosome that is deleted for most patients [18].
But besides that, it seems that the phenotypic features tend to vary with the size of the deletion and those with larger deletions show more phenotypic features and more severe presentations (hearing loss, seizures), suggesting that it would include additional modifiers genes [19].
The genomic region containing the modifier genes for prognosis of development who described two patients: One having a 5.4 Mb deletion with independent gait, the other patient with >6.1 Mb deletions, severe Intellectual Disability (ID) and no independent gait. The critical region between 5.4 Mb and 6.1 Mb contains the KCNAB2 gene, besides of the Chromodomain Helicase DNA-binding protein 5 (CHD5), involved in chromatin remodeling and gene transcription, regulating the expression of neuronal genes. These two genes were deleted in patients 1 and 3 with the largest deletions [20].
KCNAB2 was also involved in epilepsy as well as the Gammaaminobutyric Acid (GABA) A receptor delta (GABRD) gene. In fact, two patients (Pt 2, Pt 50) with no history of epilepsy, who had a 1.8 Mb terminal deletion and a 10.0 Mb interstitial deletion respectively. Both of the deletions included neither GABRD nor KCNAB2 [21].
Seizures are seen in 58% of cases. The incidence of epilepsy was higher in the patients with severe ID (79%) than in the patients with moderate ID (50%). Thus, the severity of ID was associated with the incidence of epilepsy and the same genes may be involved in both of these neurological manifestations [22].
Seizures were noted in all patients in this study. This could be due to the deletion of GABRD and KCNAB2 in patients 1 and 3 on one hand, and the deletion of GABRD in patient 2 on the other hand. These findings were expected because the ID was more severe in patients 1 and 3 in whom both genes were deleted. 24 subjects with 1p36 deletions; nine subjects with epilepsy were deleted for the KCNAB2 gene, and 13 subjects without epilepsy were not deleted for this gene. However, two subjects who were not deleted for this gene had at least one seizure episode. Thus, KCNAB2 would be a genetic modifier to the occurrence of seizures.
1p36 deletion syndrome was described to be due to terminal deletion, interstitial deletion (10%), derivative chromosome, and complex rearrangement of chromosome 1. It comes from a de novo deletion of maternally inherited chromosome 1 in 60% of patients. In our study, patient P2 had a derivative chromosome 1 inherited from her father who has a balanced translocation that could not be seen cytogenetically [23].
Because of the paternal t (1;22), patient P2 didn’t have an isolated 1p36 monosomy, but an additional 22q13 trisomy. Trisomy of the long arm of chromosome 20 is rare. Most cases have been the result of malsegregation of parental translocations or pericentric inversions. Thus the whole phenotype usually results from a variably sized trisomy 20q and different associated monosomies or trisomies. Because of the size of the trisomy and the associated monosomies in these cases, a clear genotype phenotype correlation for trisomy 20q was not always possible until pure trisomy of 20q have been described. Nevertheless, these cases of isolated trisomy 20q were resulting from duplication of a different region compared to our patient. The features are not present in patient P2, indicating that the contribution of the 20q trisomy to the clinical phenotype might be of minor importance. As a result, findings are all compatible with the 1p36 deletion syndrome.
The molecular mechanism of this malsegregation was described. It has been suggested that repetitive DNA elements were involved in the breakpoints in various constitutional chromosomal aberrations that cause genetic disease by creating aberrant nonallelic homologous recombination between closely related repetitive DNA-sequence elements. Thus, repetitive DNA sequence elements may play an important role in generating and/or stabilizing terminal deletions of 1p36.
Conclusion
The 1p36 deletion syndrome is a common malformation syndrome that is not always clinically recognizable. The diagnosis was missed by standard cytogenetic studies in our three patients, hence the advantage of FISH. Array CGH was a significant tool for patients in whom the diagnosis was not suspected. It was also useful to define breakpoints and gene content on one side, and to find unsuspected associated trisomy on the other side.
Acknowledgments
We thank Doctors Fayrouz KALLEL and Hamdi GUEDHAMI for referring the patients and providing clinical information. We are also grateful to the parents of patients for their active participation.
Conflict of Interest
None of the authors has any conflict of interest to disclose.
References
- Heilstedt HA, Ballif BC, Howard LA, Lewis RA, Stal S, et al. (2003) Physical map of 1p36, placement of breakpoints in monosomy 1p36, and clinical characterization of the syndrome. Ame J Human Gene 72: 1200-1212.
- Shapira SK, McCaskill C, Northrup H, Spikes AS, Elder FF, et al. (1997) Chromosome 1p36 deletions: The clinical phenotype and molecular characterization of a common newly delineated syndrome. Ame J Human Gene 61: 642-650.
- Battaglia A, Hoyme HE, Dallapiccola B, Zackai E, Hudgins L, et al. (2008) Further delineation of deletion 1p36 syndrome in 60 patients: A recognizable phenotype and common cause of developmental delay and mental retardation. Pediatrics 121: 404-410.
- Hain D, Leversha M, Campbell N, Daniel A, Barr PA, et al. (1980) The ascertainment and implications of an unbalanced translocation in the neonate. Familial 1:15 translocation. J Paediat Child Health 16: 196-200.
- Heilstedt HA, Ballif BC, Howard LA, Kashork CD, Shaffer LG (2003) Population data suggest that deletions of 1p36 are a relatively common chromosome abnormality. Clin Gene 64: 310-316.
- Bahi‐Buisson N, Guttierrez‐Delicado E, Soufflet C, Rio M, Cormier Daire V, et al. (2008) Spectrum of epilepsy in terminal 1p36 deletion syndrome. Epilepsia 49: 509-515.
- Colmenares C, Heilstedt HA, Shaffer LG, Schwartz S, Berk M, et al. (2002) Loss of the ski proto-oncogene in individuals affected with 1p36 deletion syndrome is predicted by strain-dependent defects in Skimice. Nat Gene 30: 106-109.
- Rosenfeld JA, Crolla JA, Tomkins S, Bader P, Morrow B, et al. (2010) Refinement of causative genes in monosomy 1p36 through clinical and molecular cytogenetic characterization of small interstitial deletions. Ame J Med Gen 152: 1951-1959.
- Zhu X, Zhang Y, Wang J, Yang JF, Yang YF, et al. (2013) 576 kb deletion in 1p36. 33-p36. 32 containing SKI is associated with limb malformation, congenital heart disease and epilepsy. Gene 528: 352-355.
- Zaveri HP, Beck TF, Hernández-García A, Shelly KE, Montgomery T, et al. (2014) Identification of critical regions and candidate genes for cardiovascular malformations and cardiomyopathy associated with deletions of chromosome 1p36. PloS One 9: e85600.
- Gajecka M, Mackay KL, Shaffer LG (2007) Monosomy 1p36 deletion syndrome. Ame J Med Gen Sem Med Gene 145: 346-356.
- Wu YQ, Heilstedt HA, Bedell JA, May KM, Starkey DE, et al. (1999) Molecular refinement of the 1p36 deletion syndrome reveals size diversity and a preponderance of maternally derived deletions. Human Mole Gene 8: 313-321.
- Zenker M, Rittinger O, Grosse KP, Speicher MR, Kraus J, et al. (2002) Monosomy 1p36-a recently delineated, clinically recognizable syndrome. Clin Dysmorphol 11: 43-48.
- Potts RC, Zhang P, Wurster AL, Precht P, Mughal MR, et al. (2011) CHD5, a brain-specific paralog of Mi2 chromatin remodeling enzymes, regulates expression of neuronal genes. PloS One 6: e24515.
- Heilstedt HA, Burgess DL, Anderson AE, Chedrawi A, Tharp B, et al. (2001) Loss of the potassium channel β‐subunit gene, KCNAB2, is associated with epilepsy in patients with 1p36 deletion syndrome. Epilepsia 42: 1103-1111.
- Shimada S, Shimojima K, Okamoto N, Sangu N, Hirasawa K, et al. (2015) Microarray analysis of 50 patients reveals the critical chromosomal regions responsible for 1p36 deletion syndrome-related complications. Brain Develop 37: 515-526.
- Plotner PL, Smith JL, Northrup H (2002) Trisomy 20q caused by der (4) t (4; 20)(q35; q13. 1): Report of a new patient and review of the literature. Ame J Med Genetics 111: 71-75.
- Grange DK, Garcia‐Heras J, Kilani RA, Lamp S (2005) Trisomy 20q13→20qter in a girl with multiple congenital malformations and a recombinant chromosome 20 inherited from a paternal inversion (20)(p13q13. 1): Clinical report and review of the trisomy 20q phenotype. Ame J Med Gene 137: 308-312.
- Sanchez O, Mamunes P, Yunis JJ (1977) Partial trisomy 20 (20q13) and partial trisomy 21 (21pter leads to 21q21. 3). J Med Gene 14: 459-462.
- Iglesias A, Rauen KA, Albertson DG, Pinkel D, Cotter PD (2006) Duplication of distal 20q: Clinical, cytogenetic and array CGH characterization of a new case. Clin Dysmorphol 15: 19-23.
- Blanc P, Gouas L, Francannet C, Giollant M, Vago P, et al. (2008) Trisomy 20q caused by interstitial duplication 20q13. 2: Clinical report and literature review. Ame J Med Gene 146:1307-1311.
- Deininger PL, Batzer MA (1999) Alu repeats and human disease. Mol Gene Metabol 67: 183-193.
- Ballif BC, Gajecka M, Shaffer LG (2004) Monosomy 1p36 breakpoints indicate repetitive DNA sequence elements may be involved in generating and/or stabilizing some terminal deletions. Chrom Res 12: 133-141.