Research Article, J Appl Bioinforma Comput Biol Vol: 7 Issue: 1
Comparative Study of Canine and Human miRNAs and CpG Islands
Wedson Gomes da Silveira1,2*, Edilson Carlos Caritá1,3, Ana L Fachin1,4 and Mozart Marins1,4*
1Unidade de Biotecnologia, Universidade de Ribeirão Preto, Ribeirao Preto SP, Brazil
2Centro Federal de Educação Tecnologica de Minas Gerais, Varginha MG, Brazil
3Centro de Ciências Exatas, Naturais e Tecnológicas, Universidade de Ribeirão Preto, Ribeirão Preto SP, Brazil
4Escola de Medicina, Universidade de Ribeirão Preto, Ribeirao Preto SP, Brazil
*Corresponding Authors : Wedson Gomes da Silveira
Unidade de Biotecnologia, University of Ribeirão Preto, Ribeirao Preto SP, Brazil
Tel: +551636036728
E-mail: wedson@varginha.cefetmg.br
Mozart Marins
Centro de Ciências Exatas, Naturais e Tecnológicas, Universidade de Ribeirão Preto, Ribeirão Preto SP, Brazil
Tel: +551636036728
E-mail: mmarins@gmb.bio.br
Received: October 05, 2017 Accepted: January 17, 2018 Published: January 23, 2018
Citation: Silveira WG, Caritá EC, Fachin AL, Marins M (2018) Comparative Study of Canine and Human miRNAs and CpG Islands. J Appl Bioinforma Comput Biol 7:1. doi: 10.4172/2329-9533.1000143
Abstract
MicroRNA (miRNA) has been recognized as a major factor in the regulation of gene expression in mammalian species and aberrant DNA methylation of tumor suppressor miRNAs due to CpG Island. Hypermethylation is emerging as another common mechanism of cancer. Because of experimental identification of miRNAs and their targets is difficult, comparative computational analysis is a popular method to identify new miRNAs and their associated targets. In this manuscript, we have performed a comparative analysis of the canine and human genome in order to find new miRNAs associated to CpG Islands. Human and canine miRNAs and genomes were obtained from mirBase and GenBank respective llythe genomes. A total of 67 miRNAs with nearby CGI were identified in the canine genome and 178 in the human genome, with one hundred and thirteen miRNAs showing some degree of homology between both species. This analysis also permitted the identification of uncatalogued miRNAs in the canine genome. The possible existence of these miRNAs is supported by the similarity score of 75% in miRbase. Our results support the notion that miRNA landscape in the canine genome is still to reveal itself in this species which develops cancers in higher rates than humans.
Keywords: Human mirnas; Protozoa; Gemomes; GenBank
Introduction
The Caenorhabditis elegans lin-4 [1] and let-7 [2] microRNAs (miRNAs) are the founding members of a large group of small nonprotein- coding RNAs, which are 20 to 24 nucleotides in length and are found in animals, plants, and protozoa [3]. A homologue of let- 7 was the first miRNA described in mammals, but thousands more have been identified since its discovery in 2000 [2]. MicroRNAs participate in the post-transcriptional regulation of the expression of genes associated with many biological processes, including those related to carcinogenesis such as cell cycle control [4], proliferation [5], apoptosis [6], and migration and invasion [7].
Mammalian miRNAs originate in the nucleus from long primary transcripts (pri-miRNAs), which are processed by the RNase III enzyme, Drosha, to form a hairpin stem-loop structure of 60–100 nucleotides (pre-miRNAs) [8]. After transport to the cytoplasm by Exportin 5, a nuclear transport receptor [9], pre-miRNAs are processed by Dicer, another RNase III family endonuclease, to yield the mature miRNA of approximately 22 nucleotides [10]. The mature miRNA is incorporated into RNA-induced silencing complexes (RISC) [11] and guide this ribonucleoprotein complex to the target mRNAs, inducing either their degradation or translational repression [12].
The aberrant expression of miRNAs as a consequence of misexpression of the miRNA machinery composed of Dicer [13], Drosha [14] and Exportin 5 [15] has been associated with different types of cancer. An additional mechanism of miRNA regulation is becoming highly evident and involves aberrant DNA methylation of miRNA genes [16]. DNA methylation is a well-known epigenetic mechanism of gene expression regulation in mammalian genomes and involves the transfer of a methyl group from S-adenosyl-L-methionine to the C5 position of cytosine, mainly in a CpG dinucleotide context. Most CpG dinucleotides in the mammalian genome are methylated; however, clusters of CpG dinucleotides, known as CpG islands (CGI), that overlap the promoter region of approximately 60% of mammalian genes, are normally unmethylated [17]. The hypermethylation of CGI in promoter regions of tumorâsuppressor genes often leads to knockdown of their expression and is a hallmark of different types of cancer [18]. More recently, aberrant DNA methylation of tumor suppressor miRNAs due to CpG island hypermethylation is emerging as another common mechanism of cancer [19-21].
Spontaneously occurring cancer is not unique in humans. It is also one of the most important canine diseases, with one in every 3-4 dogs developing some type of cancer, an incidence twice as high as in humans. The most prevalent are tumors of the mammary gland, but melanomas, lymphomas and osteosarcomas are also common [22]. They share many features with human cancers, including homology between cancer-associated genes, cytogenetic alterations, histological appearance of cancer tissues, and response to conventional therapies [23]. Indeed, the various chemotherapeutic agents used to treat human cancers are also employed in the treatment of canine cancer. However, in both cases, doubts remain regarding the development of tumor cell resistance and the lack of target specificity can lead to undesirable side effects [24].
The rate of miRNA discovery is high. In an attempt to keep track of new miRNAs, the research community relies on the public repository for microRNA sequences (miRBase). This database has been established in 2002 and stores identified, annotated and published miRNA sequences of the scientific community. The latest miRBase release (26 June 2014) contains 28,645 precursors and 35,828 mature miRNAs from 223 species, including 1,881 human precursors that produce 2,588 mature miRNA products [25]. These numbers contrast with the 502 precursors and 453 mature miRNAs curated for the canine genome. Nevertheless, cancer-associated miRNAs have also been described in dogs [26]. In this respect, a comparative genomic hybridization study of 268 canine tumor samples, including osteosarcoma, breast cancer, leukemia and colorectal cancer, indicates that cancer-associated regions with CpG islands in the dog genome tend to contain significantly more miRNA genes than non-cancerassociated regions [27]. Based on these data and the similarities between the human and canine genomes, we performed silico comparative analysis of human and canine miRNAs deposited in miRBase to identify new canine miRNAs and cancer-associated genomic regions.
Materials and Methods
Database
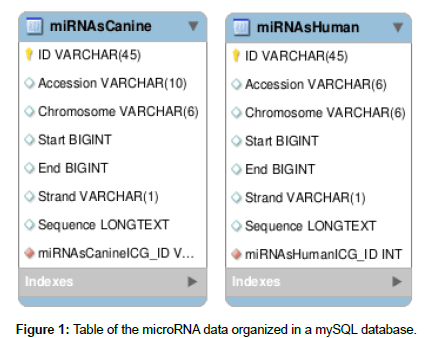
The human miRNA sequences were obtained from the miRbase repository [miRBase], and 502 canine miRNA sequences and 1,883 human miRNA sequences were catalogued. The sequences obtained were organized according to ID, name, position and chromosome using our own algorithm written in Python. After this organization, the sequences were entered in Figure 1 format into a mySQL database [oracle].
The complete canine and human genomes were obtained from the National Center for Biotechnology Information (NCBI) database. The version of the canine genome used was CanFam 3.1, updated 24 September 2013, which is available for download at: Canis lupus familiaris. The human genome used was version Hg 18, updated 15 March 2015, available at: Homo sapiens.
Computer tools
The Lift Over tool was used for the identification and analysis of similar regions in the canine and human genome. The miR Walk software (uni-heidelberg.de) was then applied to analyze and catalogue potential target genes of the miRNAs found in these regions.
Software developed
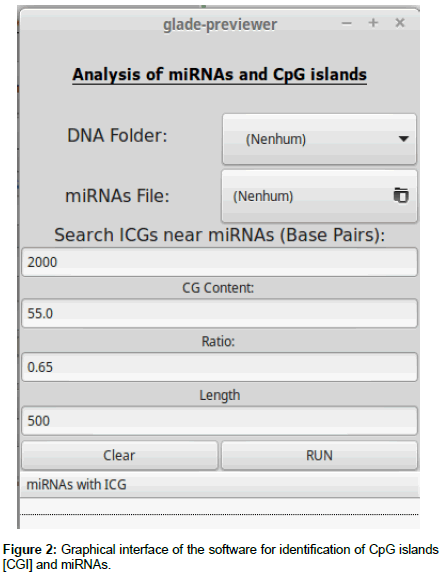
After acquisition of the data, software was developed to convert the RNA sequences into DNA and to identify CGI near miRNAs. This software was written in Python and has seven functions to obtain a list of sequences with nearby CGI as shown in Figure 2.
First, the “read_DNA” function indexes each chromosome for the subsequent queries. Two conversion functions, “rna_to_dna” and “dna_to_rna”, are used to facilitate conversions in the queries, if necessary. The “islandCpG (numberBp)” function receives as entry a value (variable qtidadePb) provided by the user, which will be the number of base pairs to be analyzed before and after the miRNA. After the function is started, a file is generated whose header informs which miRNA will have its nearby region analyzed(ID_antes), on which chromosome(Chromosome), start and final position(Start and End), the percentage of cytosine and guanine in the region(%CG), the ratio of cytosine and guanine that appear together (Ratio), and the sequence analyzed (Sequência).
Results and Discussion
Identification of miRNAs associated with CpG islands
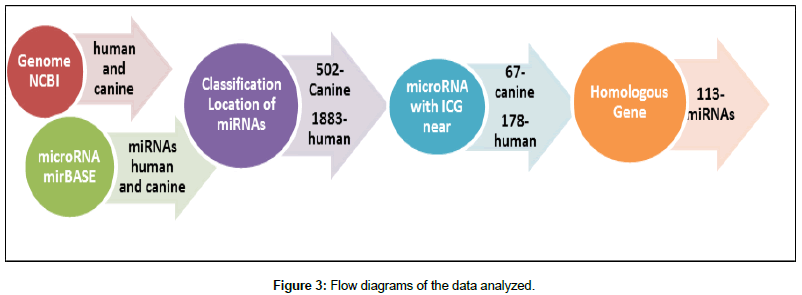
The flow diagram in Figure 3 summarizes the steps used to identify homologous miRNAs and associated CGI in the two species.
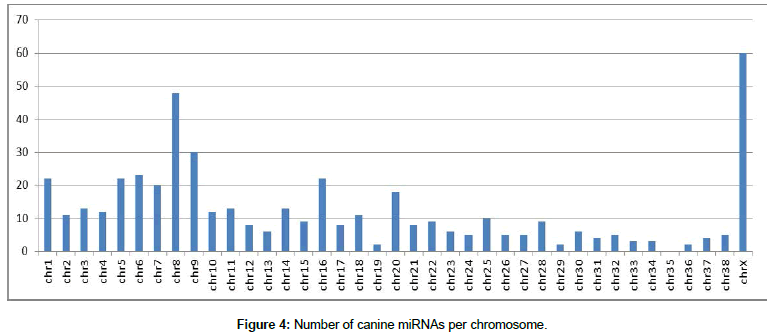
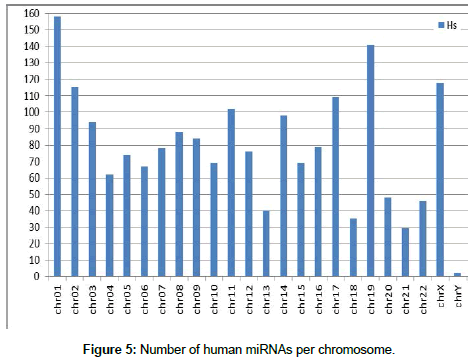
First, after organization of the miRNA data, the location of each miRNA in the canine and human genomes was determined Figures 4 and 5 show the number of miRNAs per chromosome for the canine and human genome, respectively. The miRNAs found in the genome were classified by name and position on each chromosome using our own algorithm.
With all miRNAs localized in the genome, the presence of CGI near these miRNAs was analyzed. Studies indicate a relationship between miRNAs and CGI. For example, miRNAs of the miR-200 family are associated with the regulation of cell migration/invasion. In this respect, some of these miRNA regions can predict disease and their expression is frequently associated with alterations in CpG methylation and consequently with diseases [28].
Next, using the Lift Over tool from the UCSC Genome Bioinformatics Site (genome.ucsc), the corresponding canine homologous regions comprising the miRNA sequences associated with CGI were identified in the human genome.
Following the flow chart in Figure 3, 67 miRNAs with nearby CGI were identified in the canine genome and 178 in the human genome. Analysis of miRNAs with nearby CGI in the canine and human genomes revealed annotation of 8 miRNAs shared by the two species: mir-210, mir-212, cfa-mir-1199, mir-425, mir-375, mir-132, mir- 148a, and mir-191. Based on the homology data of the two species, genes and miRNAs were investigated in the two regions. Fifty-eight miRNAs with associated CGI that were homologous in the two species were identified and are shown in Table 1.
| Human | Canine | |
|---|---|---|
| hsa-mir-4322 | S1PR2/MRPL4/ICAM1/DNMT1 | S1PR2/MRPL4/ICAM1/DNMT1 |
| hsa-mir-4750 | TBC1D17/AKT1S1/IL4I1/ATF5 | TBC1D17/AKT1S1/IL4I1/ATF5 |
| hsa-mir-760 | DNTTIP2 | DNTTIP2 |
| hsa-mir-1538 | NFAT5 | NFAT5 |
| hsa-mir-4281 | NPM1/RPL10P8/FGF18 | NPM1/RPL26L1/FGF18 |
Table 1: Genes in the homologous region shared between species.
Target genes of the miRNAs identified
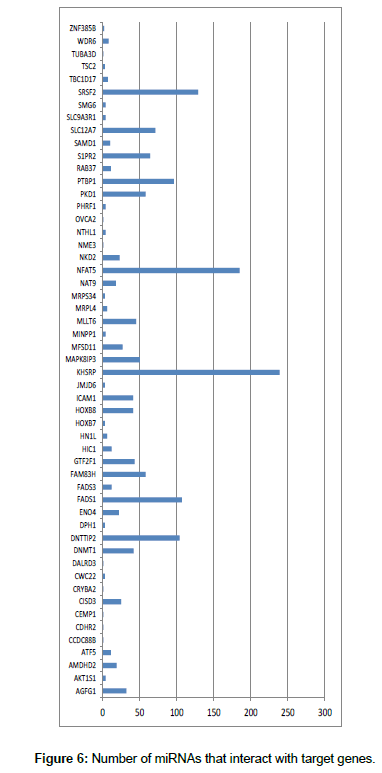
Using themiRWalk software [uni-heidelberg.mirwalk2], the interaction between the miRNAs found and their potential target genes was analyzed. The results showed that each gene is recognized by various miRNAs (Figure 5).
The genes shown in Figure 5 are located near the miRNAs and CGI. These genes have been described as miRNA target genes and are related to cancer. For example, mir-212, mir-196a-1 and mir-375 are associated with pancreatic cancer [29]. Other studies reported miir-191 to be linked to ovarian cancer. Through the relationship of miRNA and TNF-α with DAPK1 gene, it has been shown that it may contribute to the malignant tumor transformation.
The genes shown in Figure 6 were investigated in COSMIC, a database that associates the main genes with mutations and with different types of cancer. We thus obtained the miRNAs with their target genes and the genes that are directly related to cancer. Three genes directly related to mutations and cancer was identified: SRSF2, TSC2, and MLLT6. According to [30], dysregulation of SRSF2 gene expression is related to cancer. The authors confirmed the existence of abundant alternative splicing, with the SRSF2 gene being one of the main genes that contribute to the development of cancer. The TSC2gene, together with TSC1 (tuberous sclerosis complex), is linked to the development of benign tumors and some types of lesions and inflammation. In a small number of cases, these genes have also been associated with malignant tumors and cancer [31]. The MLLT6 protein is associated to oncogenic growth of cells by possibly targeting its histone modifier complex to key cancer target genes. Although its mechanism of action is not fully understood, MLLT6 is somehow linked to the development of cancer [32].
In addition to the 8 miRNAs described in the two species in the same homologous region, 5 regions were similar in the human and canine genomes, i.e., that contained the same genes, were found (Table 1). Despite great similarity, annotations of miRNAs in these regions only exist for the human genome, indicating the presence of miRNAs that have not yet been catalogued or annotated in the canine genome.
The sequences were submitted to miRbase and cluster analysis revealed a score higher than 75% for all regions. This score indicates close similarity to miRNAs already existing in the database, further supporting the existence of still uncatalogued miRNAs in these regions of the canine genome. As can be seen in Table 2, similar miRNAs were found in other species, with a similarity rate higher than 70%.
| Region studied | Association with similar miRNA | Species | Score |
|---|---|---|---|
| S1PR2/MRPL4/ICAM1/DNMT1 | dps-mir-2527 | Drosophila pseudoobscura | 77 |
| TBC1D17/AKT1S1/IL4I1/ATF5 | hsa-mir-6738 | Homo sapiens | 78 |
| DNTTIP2 | eca-mir-1302 | Equuscaballus | 91 |
| NFAT5 | hhi-miR-723 | Hippoglossushippoglossus | 76 |
| NPM1/RPL26L1/FGF18 | rno-miR-346 | Rattusnorvegicus | 78 |
Table 2: Similarity of canine sequences with existing miRNAs.
Conclusion
The analysis of all canine and human miRNAs together with nearby CGI allows obtaining a more accurate view of which miRNAs and their target genes are associated with diseases. In silico prediction is an important approach to the discovery and analysis of miRNAs. In this study using a computer-based approach for the comparative analysis of the human and canine genome, we identified 113 homologous miRNAs and associated CGI. This analysis also permitted the identification of uncatalogued miRNAs in the canine genome. The possible existence of these miRNAs is supported by the similarity score of 75% in miRbase. However, further experimental studies are necessary to confirm the existence of these miRNAs.
References
- Lee Rosalind, Feinbaum Rhonda, Victor Ambrost (1993) The C. elegansHeterochronic Gene lin-4 Encodes Small RNAs with Antisense Complementarity to &II-14. Cell 75: 843-854.
- Reinhart B, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, et.al. (2000) The 21 nucleotide let-7 RNA regulates C. elegans. Nature 403: 901-906.
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281-297.
- P. Bhattacharyya, Nitai Das, Eashita Bucha, Sudha Das, Srijit Choudhury, et al. (2016) Regulation of Cell Cycle Associated Genes by microRNA and Transcription Factor. MicroRNA 5: 180-200.
- Ziling Fang, Ling Zhang, Quan Liao, Yi Wang, Feng Yu, et al. (2017) Regulation of TRIM24 by miR-511 modulates cell proliferation in gastric cancer. Cancer Res 36: 17
- Ge H, Zou D, Wang Y, Jiang H, Wang L (2017) MicroRNA-377 DownregulatesBcl-xL and Increases Apoptosis in Hepatocellular Carcinoma Cells. Oncol Res 25: 29-34.
- Zhijun Z, Jingkang H (2017) MicroRNA-520e suppresses non-small-cell lung cancer cell growth by targeting Zbtb7a-mediated Wnt signaling pathway. Biochem Biophys Res Commun 486: 49-56.
- Lee Y, Jeon K, Lee JT, Kim S, Kim VN (2002) MicroRNA maturation: Stepwise processing and subcellular localization. EMBO J 21: 4663-4670.
- Yi Rui (2003) Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Nature 415-419.
- Park JE, Heo I, Tian Y, Simanshu DK, Chang H, et al. (2011) Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature 475: 201-205.
- Kawamata T, Tomari Y (2010) Making RISC. Trends Biochem Sci 35: 368-376.
- Iwakawa HQ, Tomari Y (2015) The Functions of MicroRNAs: mRNA Decay and Translational Repression. Trends Cell Biol 25: 651-665.
- Erler P, Keutgen XM, Crowley MJ, Zetoune T, Kundel A, et al. (2014) Dicer expression and microRNA dysregulation associate with aggressive features in thyroid cancer. Surgery 1342-1350.
- Merritt WM, Yvonne GL, Liz YH, Aparna AK, Whitney AS (2008) Dicer, Drosha, and outcomes in patients with ovarian cancer. N Engl J Med 359: 2641-2650
- Shigeyasu K, Yoshinaga O, Shusuke T, Richard B, Ajay G (2017) Exportin-5 Functions as an Oncogene and a Potential Therapeutic Target in Colorectal Cancer. Clin Cancer Res 23:1312-1322
- Suzuki Hiromu, Reo Maruyama, Eiichiro Yamamoto, Masahiro Kai (2013) Epigenetic alteration and microRNA dysregulation in cancer. Front Genet 4: 258.
- Li Shen, Chun-Xiao Song, Chuan He, Yi Zhang (2014) Mechanism and Function of Oxidative Reversal of DNA and RNA Methylation. Annual Review of Biochem 83: 585-614.
- Salvi S, Gurioli G, De Giorgi U, Conteduca V, Tedaldi G, et al. (2016) Cell-free DNA as a diagnostic marker for cancer: current insights. Onco Targets Ther 9: 6549-6559
- João Ramalho-Carvalho, Inês Graça, Antonio Gomez, Jorge Oliveira, Rui Henrique (2017) Downregulation of miR-130b~301b cluster is mediated by aberrant promoter methylation and impairs cellular senescence in prostate cancer. J Hematol Oncol 10: 43.
- Yuan Yang, Linghao Zhao, Bo Huang, Guojun Hou, Beibei Zhou, et al. (2017) A New Approach to Evaluating Aberrant DNA Methylation Profiles in Hepatocellular Carcinoma as Potential Biomarkers. Sci Rep 7: 46533.
- Amaia L, George AC, Alberto V, Santiago R, Montserrat SC, et al. (2008) A microRNA DNA methylation signature for human cancer metastasis. Cancer Research Center 105: 13556-13561.
- Dobson Jane M (2013) Breed-Predispositions to Cancer in Pedigree Dogs. ISRN Veterinary Science.
- Paoloni Melissa and Khanna Chand (2008) Translation of new cancer treatments from pet dogs to humans. Nat Rev Cancer 8:147-156
- MacDonald Valerie (2009) Chemotherapy: Managing side effects and safe handling. Can Vet 50: 665-668.
- Kozomara, Ana and Griffiths-Jones, Sam. (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42: D68–D73.
- Wagner Siegfried, Saskia Willenbrock, Ingo Nolte, Hugo Murua Escobar (2013) Comparison of non-coding RNAs in human and canine cancer. Front Genet 4: 46.
- Mohamad Zamani-Ahmadmahmudi, Sina Aghasharif, Keyhan Ilbeigi (2016) Prognostic efficacy of the human B-cell lymphoma prognostic genes in predicting disease-free survival [DFS] in the canine counterpart. BMC Vet Res 13: 17.
- Long MD, Smiraglia DJ, Campbell MJ (2017) The Genomic Impact of DNA CpG Methylation on Gene Expression; Relationships in Prostate Cancer. Biomolecules 7: 15.
- Lee YS, Dutta A (2009) MicroRNAs in cancer. Annu Rev Pathol 4:199-227
- Luo C, Cheng Y, Liu Y, Chen L, Liu L, et al. (2017) SRSF2 regulates alternative splicing to drive hepatocellular carcinoma development. Cancer Res 77:1168-1178.
- Katie RM, Wanding Z, Megan JB, Juliann S, Kit SA, et al. (2017) The genomic landscape of tuberous sclerosis complex. Nature Research Journal.
- Slobodan B, Peter J, Evan H, Wen-HL, Brice EK (2013) RNAi screens in mice identify physiological regulators of oncogenic growth. Nature 501: 185-190.