Research Article, J Genet Disor Genet Rep Vol: 11 Issue: 6
Contribution of Functional Genes to the Anthropogenetic Characterization of Two Moroccan Berber Populations from Souss and Ouarzazate: Apolipoproteins E and CI Genes
FE Anaibar1*, M Kandil1, N Harich1, P Moral2 and E Esteban2
1Department of Sciences, University of Chouaib Doukkali, El Jadi, Maroc
2Department of Biology, University of Barcelona, Barcelona, Spain
*Corresponding Author: F E Anaibar Department of Sciences, University of Chouaib Doukkali, El Jadi, Maroc, Tel: 212 665152028; E-mail: anaibar.fe@gmail.com
Received: 13 December 2020, Manuscript No. JGDGR-20-23291; Editor assigned: 16 December 2020, PreQC No. JGDGR-20-23291 (PQ); Reviewed: 30 December 2020, QC No. JGDGR-20-23291; Revised: 30 July 2022, QI No. JGDGR-20-23291; Manuscript No. JGDGR-20-23291 (R); Published: 29 August 2022, DOI:10.4172/2327-5790.1000032
Citation: Anaibar FE, Kandil M, Harich N, Moral P, Esteban E (2022) Contribution of Functional Genes to the Anthropogenetic Characterization of Two Moroccan Berber Populations from Souss and Ouarzazate: Apolipoproteins E and CI Genes. J Genet Disor Genet Rep 11:6.
Abstract
Introduction: Genetic studies of human populations have as main purpose, the characterization of population allelic frequencies and the analysis of the diversity to assess their affinity or differentiation from other populations, according to the ethnicity, region or language groups. The use of susceptibility markers to characterize some human diseases in epidemiological studies could be of interest in the anthropological characterization of human populations. Thus, our results can constitute an interesting source of data and a supplement of information for the reconstruction of the biological history of the analysed samples.
Methods: In this context our study focused on the analysis of molecular polymorphisms of two apolipoprotein genes: ApoE and ApoCI, in two moroccan berber-speaking populations. The aim of this study is to predict genetic predisposition to develop cardiovascular disease and to study genetic affinities with other Moroccan and Mediterranean populations.
Results and discussion: The Berber speaking populations from the regions of ouarzazate and souss have been characterized for the first time by the two studied markers. The comparisons of allelic frequencies allowed us to dress a profile of studied populations’ affinities together with other Moroccan and Mediterranean populations. This profile allowed to highlight patterns of differentiation similar to others obtained for these two populations with other types of markers supposed to be neutral.
Keywords: Population; Apolipoproteins; Functional genes;
Anthropogenetic
Introduction
The genetic markers used in human population genetics studies are of different types: Conventional markers and molecular markers that can have both applications in population and epidemiological genetic studies.
Cardio Vascular Disease (CVD) is one of the leading causes of death in industrialized countries. In Europe, the Mediterranean region is characterized by low frequencies of CVD compared with other regions, this low level is related to protective environmental factors and genetic background. In the North African region, Morocco seems to have the same tendency. Today, diseases associated with lipid accumulation in the human body, such as obesity or atherosclerosis represent principal factors promoting heart disease and are becoming increasingly important in health issues. These lipid accumulations are the result of an unbalanced diet, coupled to the sedentary life, the lack of physical activity, stress, alcohol, tobacco [1].
Another important cause of these accumulations is the genetic predisposition correlating with the variations affecting candidate genes. Apoliprotein genes have been involved in a direct relationship with plasma lipoprotein levels, especially under some allelic combinations. Numerous epidemiological studies have shown that when these levels are altered, they represent a major risk factor for cardiovascular disease. This work aims to provide additional data by the analysis of molecular polymorphisms on apolipoprotein gene cluster ApoE-CI, in order to complete the analysis of two Moroccan populations from ouarzazate and souss Berber-speaking regions.
The main objectives of the genetic characterization of the sampled Moroccan populations are the determination of the allelic frequencies of ApoE and ApoCI genes, the comparison of their frequencies distribution with other Moroccan and Mediterranean populations, the analysis of their genetic affinity together and with the other populations and finally the exploration of possible genetic predisposition to develop cardiovascular disease [2].
General characteristics of lipoproteins
Lipids are insoluble in water and are transported in the plasma only when they are associated, with one or more specific proteins called apolipoproteins or apoproteins on their surface. This association ensures also the stability of the macromolecule. The main physiological role of circulating lipoproteins is the transport and the distribution of exogenous and endogenous lipids between the different tissues involved in their metabolism. Plasma lipoproteins have common structural features; they comprise several different families classified according to their composition, metabolism and physiological role [3,4].
General characteristics of apolipoprotein
The apolipoproteins form the protein part of lipoproteins; they play a fundamental role in lipid blood transportation and in their metabolism. They can also have other specific functions such as playing the role of cofactor like the plasma activator of many enzymes and serving as ligands for cellular receptors of lipoproteins. Apolipoproteins form a large protein family with similar functions, structures and sequences.
The gene of ApoCI is located on 4.3 kb or 5.3 kb downstream of the ApoE gene. It is expressed primarily in the liver and in lower levels in the lung, skin, testis and spleen. It encodes a protein of 57 amino acids [5].
ApoCI inhibits the purification of plasma triglycerid-rich lipoproteins (VLDL and chylomicrons) by hepatic receptors. This inhibition is accomplished by moving the ApoE on the surface of these lipoproteins or by altering its conformation, which prolongs their residence time in plasma and promotes the conversion of VLDL into IDL and LDL. The ApoCI is an activator of LCAT, an enzyme that esterified cholesterol, but also a main inhibitor of CETP that promotes the exchange of cholesterol esters from HDL to VLDL.
The ApoE gene is the most studied from the large family of apolipoproteins. It located at the centromeric end of a long cluster of apolipoprotein genes coding for the group of the apolipoproteins E, CI and CII. The chromosomal location of ApoE is the q13.2 region on chromosome 19, along a distance of 3.6 kb consisting of three exons that encodes for a mature protein of 299 amino acids with a molecular weight of 34 kDa [6].
Materials and Methods
Subjects
Our study involved 200 indivuals, 100 from each region: Ouarzazate and souss. Individuals’ selection was carried out first according to the origin, the speaking language and the information obtained from their familial genealogy (parents and grandparents born in the same region) All the individuals are unrelated, apparently healthy and have signed an informed consent [7].
Blood samples were obtained by venipuncture from consentent volontaires by the experimented medical staff from each region. The anticoagulant of the sampling tubes was EDTA and the whole blood was fractioned after settling overnight at 4°C.
DNA extraction
DNA was extracted from the leucocytal fraction using the Promega kit and amplified by Polymerase Chain Reaction (PCR) in a DNA thermal cycler using oligonucleotide primers and amplification reaction conditions as described for ApoCI.
Digestion
The PCR products of ApoCI and ApoE were digested with HincII and HhaI enzymes respectively.
Electrophoresis
The digestion products were then separated by 2% agarose gel for ApoCI and 10% Acrylamide Gel electrophoresis for ApoE. The restriction of the amplification products of APOE was performed by restriction enzyme HhaI.
Statistical analysis
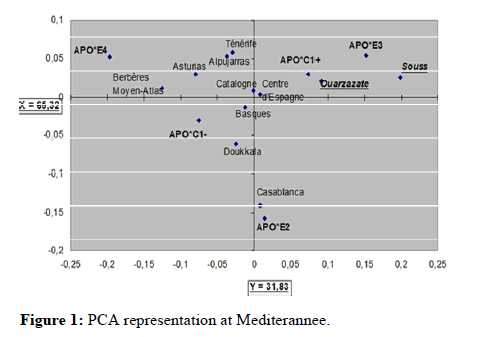
To calculate the allele frequencies of the two polymorphic systems ApoE and ApoCI analyzed in this study, we used the Biosys-1 computer program. This program verifies also the hardy-Weinberg equilibrium using a χ2 test, giving the probability that chance alone is the source of discrepancies between the observed and theoretical frequecies, and also the heterozygosity of each system. Comparisons of the allelic frequencies distributions of each marker in both populations Ouarzazate and Souss were done using available literature data for Moroccans and Mediterranean populations. These comparisons were also performed using the program Biosys-1 using the χ2 test to check whether the differences, between the allelic frequencies distributions, are significant or not. Principal Component Analysis (PCA) was performed using the R-matrix to visualize the genetic relationships among populations based on the raw data of allele frequencies (Figure 1) [8,9].
Results and Discussion
The heterozygosity for ApoCI gene was 27.7% and 25.1% among respectively the populations of Ouarzazate and Souss. This analysis also shows that the most common allele of ApoCI in both populations is H- with values of 81.2% and 78.9% respectively in Ouarzazate and Souss as it is the case of all Mediterranean populations (Table 1).
| Allele frequencies | |||
|---|---|---|---|
| Ouarzazate | Souss | ||
| ApoE (HhaI) | |||
| E2 | 0.045 | E2 | 0.045 |
| E3 | 0.9 | E3 | 0.949 |
| E4 | 0.055 | E4 | 0.005 |
| N | 100 | N | 99 |
| H | 0.186 | H | 0.097 |
| ApoCI (HincII) | |||
| H+ | 0.188 | H+ | 0.211 |
| H- | 0.812 | H- | 0.789 |
| N | 40 | N | 38 |
| H | 0.277 | H | 0.251 |
Table 1: Allele frequencies ApoE and ApoCI systems in both polpulation.
Allele frequencies and heterozygosity
ApoE genotypic frequencies the ε3ε3 genotype is the most common, hence the high frequency of the ε3 allele in both populations with a value of 90% for ouarzazate and 94.9% for souss, populations. There is also a relatively high value of the ε2 allele (4.5% for both populations) compared to that of ε4 5.5% and 0.5% respectively. Ouarzazate population has therefore a much higher frequency of ε4 than the Souss one. Heterozygosity as a measure of the intrapopulational diversity was H=0.18 in the population of ouarzazate, and H=0.09 in the population of souss [10].
The results for the allele frequencies of ApoCI (Table 1), show that the population of Ouarzazate is in Hardy-Weinberg Equilibrium (χ2=3.11, df=1) while that of Souss is not (χ2=5.11, df=1). This imbalance could be attributed to the small sample size.
Intermediate value of the Mediterranean region, which range of variation vary between [2.1% (Tenerife)-7.9% (Turkey and France)].
The frequency of allele APO-E3 (90%) represent the new upper bound of the range of variation in the Mediterranean basin region [80.1% (France)-89.8% (Corsica)], while the APOE4 frequency in this population can be considered as one of the lowest values recorded in the Mediterranean region.
Inter-populational comparisons: Comparisons are made with the bibliographic data available for each marker.
ApoE allelic distribution
For ouarzazate’s alleles distribution of the ApoE gene frequency of APO-E2 allele among the Berbers of ouarzazate (4.5%) is equal to that of the Middle Atlas Berbers, it can be classified as an Considering the % of non-significant differences (%NSD), ouarzazate’s distribution recorded a value of 66.7% (2/3) with the Moroccan populations (the only difference was recorded with the Berbers of the Middle Atlas), and a value of 82.35 (14/17) with the other Mediteranean peoples of the north shore of the Mediterranean (in this case, significant differences were recorded with populations of Spain (two populations) and France (one population) (Tables 2 and 3).
| N | APO-E2 | APO-E3 | APO-E4 | ||
|---|---|---|---|---|---|
| Berbers of Ouarzazate | 100 | 0.045 | 0.9 | 0.055 | --- |
| Morocco | |||||
| Berbers of Middle Atlas | 121 | 0.045 | 0.798 | 0.157 | 11.65 |
| Arabs Doukkla | 101 | 0.074 | 0.837 | 0.089 | 3.53NS |
| Casablanca (general popl) | 50 | 0.11 | 0.83 | 0.06 | 4.63NS |
| Spain | |||||
| Basques | 111 | 0.05 | 0.865 | 0.086 | 1.58NS |
| Catalonia | 88 | 0.045 | 0.864 | 0.091 | 1.82NS |
| Asturias | 96 | 0.036 | 0.828 | 0.135 | 7.47 |
| Centre of Spain | 120 | 0.046 | 0.871 | 0.083 | 1.35NS |
| Alpujarras | 99 | 0.025 | 0.859 | 0.116 | 5.65NS |
| Tenerife | 72 | 0.021 | 0.868 | 0.111 | 4.86NS |
| Spain (general pop) | 141 | 0.042 | 0.831 | 0.127 | 7.02 |
| France | |||||
| France (general pop) | 498 | 0.079 | 0.801 | 0.12 | 11.17 |
| Corse | 178 | 0.041 | 0.898 | 0.061 | 0.12NS |
| Italy | |||||
| Italy | 417 | 0.066 | 0.851 | 0.083 | 3.19NS |
| Northern Italiy | 352 | 0.054 | 0.857 | 0.098 | 3.60NS |
| Centre of Italy | 365 | 0.072 | 0.847 | 0.081 | 3.71NS |
| Southern Italy | 367 | 0.056 | 0.858 | 0.085 | 2.41NS |
| Sicile | 159 | 0.064 | 0.829 | 0.107 | 5.21NS |
| Sardaigne | 188 | 0.065 | 0.826 | 0.109 | 5.84NS |
| Greece | 335 | 0.054 | 0.876 | 0.07 | 0.86NS |
| Turkey | 8366 | 0.079 | 0.86 | 0.061 | 3.39NS |
Table 2: Interpopulational comparisons of the APOE alleles distribution for Ouarzazate Berbers in the Mediterranean region.
| N | APO-E2 | APO-E3 | APO-E4 | ||
|---|---|---|---|---|---|
| Berbers of Souss | 99 | 0.045 | 0.949 | 0.005 | |
| Morocco | |||||
| Berbers of Middle Atlas | 121 | 0.045 | 0.798 | 0.157 | 31.28 |
| Arabs Doukkla | 101 | 0.074 | 0.837 | 0.089 | 17.68 |
| Casablanca ( general popl) | 50 | 0.11 | 0.83 | 0.06 | 13.71 |
| Spain | |||||
| Basques | 111 | 0.05 | 0.865 | 0.086 | 15.12 |
| Catalonia | 88 | 0.045 | 0.864 | 0.091 | 15.87 |
| Asturias | 96 | 0.036 | 0.828 | 0.135 | 25.74 |
| Centre of Spain | 120 | 0.046 | 0.871 | 0.083 | 14.61 |
| Alpujarras | 99 | 0.025 | 0.859 | 0.116 | 22.21 |
| Tenerife | 72 | 0.021 | 0.868 | 0.111 | 20.91 |
| Spain (general pop) | 141 | 0.042 | 0.831 | 0.127 | 24.6 |
| France | |||||
| France (general pop) | 498 | 0.079 | 0.801 | 0.12 | 28.68 |
| Corse | 178 | 0.041 | 0.898 | 0.061 | 10.26 |
| Italy | |||||
| Italy | 417 | 0.066 | 0.851 | 0.083 | 17.09 |
| Northern Italiy | 352 | 0.054 | 0.857 | 0.098 | 18.74 |
| Centre of Italy | 365 | 0.072 | 0.847 | 0.081 | 17.45 |
| Southern Italy | 367 | 0.056 | 0.858 | 0.085 | 16.34 |
| Sicile | 159 | 0.064 | 0.829 | 0.107 | 21.31 |
| Sardaigne | 188 | 0.065 | 0.826 | 0.109 | 22.16 |
| Greece | 335 | 0.054 | 0.876 | 0.07 | 12.84 |
| Turkey | 8366 | 0.079 | 0.86 | 0.061 | 14.78 |
Table 3: Interpopulational comparisons of APOE allelic distribution of Souss Berbers in the Mediterranean.
Concerning ApoE allelic distribution in the Souss’ population comparisons with the same list of populations from the Mediterranean region, shows that the APOE2 in Berber from Souss (4.5%) is equal to that of the other Berbers from Ouarzazate and Middle Atlas. This frequency is lower than that of two Moroccan Arabic Moroccan population (Arabs from Casablanca and Doukkala 7.4% and 11% respectively), and elongs to the range of variation of the Mediterranean region. The frequency of allele APOE3 (94.9%), more higher than that of ouarzazate berbers can be considered as the new bound of the range of variation of the Mediterranean basin [80.1% (France)-90% (ouarzazate)]. The frequency of the APOE4 allele (0.5%) represent the lowest value of the entire Mediterranean basin. It would be the new lower bound of the range of variation in this region [6.1% (Corsica)-15.7% (Middle Atlas Berber)].
Comparisons results showed that the NSD% is 0% with both Moroccan populations and with the other populations from the northern shore of the Mediterranean Sea. These results shows so the uniqueness of the distribution of this system in the Souss population, especially for allelic frequencies of APOE3 and APOE4.
APOCI allelic distribution
Tables 4 and 5 comparisons between the allelic frequencies distributions of the ApoCI gene in Ouarzazate and in Souss respectively, both with those of Moroccan and those of North Mediterranean populations. The distribution of allele frequencies of ApoCI in ouarzazate did not present significant differences compared to the populations from the Mediterranean (DNS%=100%). While that souss has submitted only one significant difference with the population of Sardinia (%DNS=91%). These results confirm the homogeneity of the allelic frequencies of this system in Morocco and in the Mediterranean [11].
| N | APOCI-H+ | APOCI-H- | ||
|---|---|---|---|---|
| Berbers of Ouarzazate | 40 | 0.188 | 0.813 | --- |
| Morocoo | ||||
| Berbers of Middle Atlas | 120 | 0.15 | 0.85 | 0.63 NS |
| Arabs Doukkala | 101 | 0.139 | 0.861 | 1.06NS |
| Spain | ||||
| Catalonia | 88 | 0.153 | 0.847 | 0.47 NS |
| Basques | 111 | 0.126 | 0.874 | 1.81 NS |
| Asturias | 90 | 0.15 | 0.85 | 0.58 NS |
| Centre of Spain | 120 | 0.146 | 0.854 | 0.79 NS |
| Alpujarras | 100 | 0.15 | 0.85 | 0.60 NS |
| Tenerife | 74 | 0.142 | 0.858 | 0.81 NS |
| Italy | ||||
| Sicile | 159 | 0.132 | 0.868 | 1.60 NS |
| Sardaigne | 188 | 0.121 | 0.879 | 2.66 NS |
| France | ||||
| Corse | 178 | 0.126 | 0.874 | 2.05 NS |
Table 4: Comparisons interpopulationnelles of APOCI system of Ouarzazate Berbers in the Mediterranean.
| N | APOCI-H+ | APOCI-H- | ||
|---|---|---|---|---|
| Berbers of Ouarzazate | 40 | 0.188 | 0.813 | --- |
| Morocoo | ||||
| Berbers of Middle Atlas | 120 | 0.15 | 0.85 | 0.63 NS |
| Arabs Doukkala | 101 | 0.139 | 0.861 | 1.06NS |
| Spain | ||||
| Catalonia | 88 | 0.153 | 0.847 | 0.47 NS |
| Basques | 111 | 0.126 | 0.874 | 1.81 NS |
| Asturias | 90 | 0.15 | 0.85 | 0.58 NS |
| Centre of Spain | 120 | 0.146 | 0.854 | 0.79 NS |
| Alpujarras | 100 | 0.15 | 0.85 | 0.60 NS |
| Tenerife | 74 | 0.142 | 0.858 | 0.81 NS |
| Italy | ||||
| Sicile | 159 | 0.132 | 0.868 | 1.60 NS |
| Sardaigne | 188 | 0.121 | 0.879 | 2.66 NS |
| France | ||||
| Corse | 178 | 0.126 | 0.874 | 2.05 NS |
Table 5: Comparisons inter populationnelles of APOCI system of Souss Berbers in the Mediterranean.
APOCIH frequency in souss berbers (21.1%) remains slightly higher than that of the Berbers of Ouarzazate (18.8%) and than the Middle Atlas Berbers (15%). The population of the Souss presents so the highest value of this allele encountered so far in the Mediterranean which values range is [12.1% (Sardinia), 15.3% (Catalonia)]. Similarly, the frequency encountred in ouarzazate can be considered as the second highest value in the Mediterranean region.
Principal component analysis
Coefficients of relationship: The coefficients of kinship between populations obtained in the analysis in the Mediterranean between Moroccan populations only souss and ouarzazate berbers showed a positive value (Table 6).
| 1- | 2- | 3- | 4- | 5- | 6- | 7- | 8- | 9- | 10- | 11- | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1-Ouarzazate | 91 | 190 | -102 | -35 | -21 | -23 | -23 | 1 | 5 | -20 | -62 |
| 2- Souss | 406 | -238 | -64 | -19 | -60 | -35 | 1 | 15 | -47 | -147 | |
| 3-BMA | 178 | 21 | -26 | 50 | -7 | 0 | -20 | 33 | 112 | ||
| 4-Douk | 43 | 83 | -23 | 12 | -5 | -4 | -27 | 0 | |||
| 5-Casa | 201 | -79 | 15 | -13 | -5 | -87 | -49 | ||||
| 6-ALPU | 43 | 0 | 5 | 0 | 44 | 44 | |||||
| 7-BASQ | 25 | 1 | 8 | 8 | -3 | ||||||
| 8-CATA | 1 | 1 | 7 | 2 | |||||||
| 9-C.SPAIN | 5 | 5 | -10 | ||||||||
| 10-TENE | 50 | 36 | |||||||||
| 11-ASTU | 76 | ||||||||||
Table 6: Kinship coefficients (10-4) and participation in the total diversity of Mediterranean populations according to APOE and APOCI.
Regarding the contribution to the total diversity of the PCA analysis, souss population brings the largest contribution (406.10-4), followed by Casablanca with (201.10-4), and by the Berbers from Middle Atlas (178.10-4). The lowest contribution has been recorded by the Doukkala population (43.10-4) [12]. PCA representation in the Mediterranean shows that the first two axes represent 97.15% of the total variability. Along the first axis (65.32%), there is a clear separation between the Moroccan Berber populations (souss and ouarzazate) occupying the X part, while Middle Atlas berbers occupy the X (-) one. Moroccan Arabic-speaking population (Doukkala) and other European populations of the northern Mediterranean side occupy majoritarely X. The two Moroccan Berber populations souss and ouarzazate are characterized mainly by high frequency of the APO-E3 allele. The other Berber population of Morocco from Middle Atlas is characterised by a high frequency of APO-E4. Morrocan Arabicspeaking populations (Doukkala and Casablanca) are characterized by high frequencies of APOE2. We can say that there is a clear separation between the populations belonging to the Moroccan Berbers groups, together and with the Arabic-speaking ones. So, the two new analysed berber populations are affine together and are heterogeneus with the other populations of the PCA analysis both at the Moroccan and at the Mediterranean scales [13].
This analysis shows a very high efficiency of these two markers in separating people from both shores of the Mediterranean and that the separating people from both shores of the Mediterranean and that the alleles that contribute most to this segregation are those of the APOE system.
Conclusion
The molecular markers located at functional genes used in this study allowed the genetic characterization of two Moroccan Berberspeaking populations: Ouarzazate and souss, and allowed us to show some special features in each one. For the APOE system, the two populations shows the highest frequency of APO-E3 allele in the Mediterranean and for the APOCI, they shows also the highest values of the APOCI-H+ in this region. The ApoE-CI cluster is fast becoming a marker of choice in the study of Mediterranean populations’ differenciation, especially between Moroccan Berbers together and with Arabic groups. This is marked at the PCA analysis by a clear separation between the Berber and Arab populations.
On the other hand, the PCA analysis and the χ2 comparisons, showed the APOE genetic variability includes that of all the Mediterranean populations. To determine the correlation between the incidence of cardiovascular disease and the heterogeneity of genotypes, it is planned to continue our study of the polymorphism of the cluster ApoE-CI-CII on a larger sample, to extend the study of other healthy populations and samples of patients suffering cardiovascular diseases, with an increment of the number of markers involved in this pathology.
References
- Bailleul S, Couderc R, Landais V, Lefevre G, Raichvarg D, et al. (1993) Direct phenotyping of human apolipoprotein E in plasma: Application to population frequency distribution in Paris. Hum Hered 43: 159-165
- Bolanos-Garcia VM, Miguel RN (2003) On the structure and function of apolipoproteins: More than a family of lipid-binding proteins. Prog Biophys Mol Biol 83: 47-68.
- Corbo RM, Scacchi R, Mureddu L, Mulas G, Alfano G (1995) Apolipoprotein E polymorphism in Italy investigated in native plasma by a simple polyacrylamide gel isoelectric focusing technique. Comparison with frequency data of other European populations. Ann Hum Genet 59: 197-209.
- Das M, Gursky O (2015) Amyloid-forming properties of human apolipoproteins: Sequence analyses and structural insights. Adv Exp Med Biol 855: 175-211.
- Gerdes LU, Klausen IC, Sihm I, Faergeman O, Vogler GP (1992) Apolipoprotein E polymorphism in a Danish population compared to findings in 45 other study populations around the world. Genet Epidemiol 9: 155-167.
- Geisel J, Weißhaar C, Oette K (1996) Anavall polymorphism in the human apolipoprotein C-II gene. Clin Genet 49: 163.
- Harich N, Esteban E, López‐Alomar A, Chafik A, Moral P (2002) Apolipoprotein molecular variation in Moroccan Berbers: pentanucleotide (TTTTA) n repeat in the LPA gene and APOE‐C1‐C2 gene cluster. Clin genet 62: 240-244.
- Holvoet P (2012) Stress in obesity and associated metabolic and cardiovascular disorders. Scientifica.
- James RW, Boemi M, Giansanti R, Fumelli P, Pometta D (1993) Underexpression of the apolipoprotein E4 isoform in an Italian population. Arterioscler Thromb 13: 1456-1459.
- Lund-Katz S, Phillips MC (2010) High density lipoprotein structure-function and role in reverse cholesterol transport. Subcell Biochem 51: 183-227.
- Piepoli MF, Hoes AW, Agewall S, Albus C, Brotons C, et al. (2016) European Guidelines on cardiovascular disease prevention in clinical practice. Polish Heart J 74: 821-936.
- Swofford DL, Selander RB (1989) BIOSYS-1: A computer program for the analysis of allelic variation in population genetics and biochemical systematics, release 1.7. Illinois Natural History Survey Champaign.
- Tsai MS, Tangalos EG, Petersen RC, Smith GE, Schaid DJ, et al. (1994) Apolipoprotein E: Risk factor for Alzheimer disease. Am J Hum Genet 54: 643.