Research Article, J Appl Bioinforma Comput Biol Vol: 7 Issue: 1
CRISPR Cas9 Nuclease Recruitment for Targeted Mutagenesis using Oligo SgRNA in Soybean (Glycine Max L)
Nooral Amin1, Naveed Ahmad2, Wu Nan1, Ma Tong1, Pu Xiumin1, Wang Nan1, Xiaoxue Bo1, Rahat Sharif3, Asad Rehman1, Asad Riaz1 and Wang Piwu1*
1College of Agronomy, Plant Biotechnology Centre, Jilin Agricultural University, Changchun 130118, Jilin, China
2College of Life Sciences, Crop biotechnology Centre, Jilin Agricultural University, Changchun 130118, Jilin, China
3College of horticulture, Northwest Agriculture, and Forestry University Yangling, Shaanxi 712100, China
*Corresponding Author : Wang Piwu
College of Agronomy, Plant Biotechnology Centre, Jilin Agricultural University, Changchun 130118, Jilin, China
Tel: 0431- 84532908
E-mail: peiwuw@163.com
Received: January 01, 2018 Accepted: February 05, 2018 Published: February 12, 2018
Citation: Amin N, Ahmad N, Nan W, Tong M, Xiumin P, et al. (2018) CRISPR Cas9 Nuclease Recruitment for Targeted Mutagenesis using Oligo SgRNA in Soybean (Glycine Max L). J Appl Bioinforma Comput Biol 7:1. doi: 10.4172/2329-9533.1000146
Abstract
The Modern era of genome engineering via targeted mutagenesis is now bringing further innovations in the field of Plant biology and biomedicines. RNA-guided endonucleases CRISPR system has been proved as most powerful and accurate tool to study the mechanism of genome screening and engineering. We report the collection of resources that can redirect and ensures the flexibility of targeted genomic alteration in eukaryotes. Single guide RNA can be recruited more precisely and efficiently to become a powerful tool in genome manipulation overcoming a broader aspect of synthetic biology. CRISPR P2 is a local tool known for the optimization of sgRNA in plant research. The entire overview of all possible single gRNA on-targeted mutagenesis in any user-provided sequence has been demonstrated along with specified PAM in soybean seed. We hope to deliver enough confidence for our readers to get started and design your own graphical genome model which could significantly improve your research.
Keywords: CRISPR Cas9 nuclease; Targeted mutagenesis; SgRNA; Cloning
Background
Soybean genome can be applied in basic as well as advanced genomic research by exploring various agricultural improvements. In recent years a new tool, the CRISPR Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats) for targeted mutagenesis revolutionized the field of genome engineering. CRISPR system is inherited from the adaptive immune system of many prokaryotes and archaea against various pathogens including viruses [1-3]. This system includes CRISPR-associated Cas9 nuclease which cleaves nucleic acid sequence and a synthetic small guide RNA which guide Cas9 complex introducing double strand break (DSB) in selected nucleotide sequence. Small guide RNA is the first 20 nucleotide sequences complementary to one strand of the target DNA (protospacer) with an -NGG motif in 3’-end (the protospacer adjacent motif, PAM) of the target site which scan a target genome for editing. After complete cycle of hybridization among single guide RNA and its desired DNA sequences the Cas9 nuclease brings conformational changes by activating its two domains causing cleavage in the target nucleotide sequence. The phenomenon has been well studied in various plants and animals which reveal the formation of hairpin structure from the 3′end of small guide RNA which actually interacts with cas-9 nuclease and direct it towards targeted mutagenesis [4].
The two known mechanisms of DNA repair system in the eukaryotic cells, non homologous end joining (NHEJ) and homologous recombination [5]. NHEJ leads to various kinds of mutation (deletion, substitution and insertion). These mutations can drastically influence the regulation and expression of target genes when it occur in the coding or promoter regions [6,7]. Homologous recombination (HR) involves the repair of a DSB (DNA Stranded Break) by homologous donor DNA which serves as a template which increases the accuracy of insertion or replacement [8]. CRISPRCas9 system is very simple, fast, more efficient and easy to design than ZFNs (Zinc Finger Nuclease) and TALENs (Transcription Activator Like Effectors Nuclease), that’s why the CRISPR system is broadly used for genome engineering and promoting emerging applications of this system [9,10]. CRISPR/Cas9 mediated transient expression system has been successfully studied in wheat and soybean protoplasts [7,11]. The CRISPR Cas9 induced transgenic lines of Arabidopsis and rice up to first generation was observed recently [12,13].
Here we demonstrate CRISPR P2.0 a downloadable, easily accessed and open online tool to recruit a small guide RNA sequence associated with CRISPR Cas9 nuclease system to tackle the possible potential sites in any given genome. We have successfully designed SgRNA based CRISPR Cas-9 vector for FAD2-2 gene yielding high oleic acid content in soybean seed.
However careful evaluation need to be considered which can considerably affect the specificity of CRISPR Cas9 system [14,15]. Our study suggested that variation in mismatch number, PAM type, snoRNA promoter; RNA scaffold and Guide sequence length influence the specificity of targeted mutagenesis. Although the CRISPR Cas9 system has been widely and successfully applied in many crop plants, there is no report on the efficacy of SgRNA taken from FAD-2 gene for the manipulation of FAD2-2 gene in Soybean.
Designing and Implementation
Searching tools for single gRNA
CRISPR-P: An open web tool which can be locally accessible through Mac OS system and windows were exploited in this report for synthetic SgRNA designing of CRISPR Cas9 system in soybean. Searching mode for on target and off target annotations in FAD2-2 gene of glycine max L. has been demonstrated. Several links to open sequences of Glycine max L. FAD2-2 microsomal omega-6 desaturase mRNA are available on several widely used databases. In order to construct synthetic SgRNA guide we extracted FASTA file of the complete Cds of FAD2-2 gene having GenBank Accession number (L43921) from NCBI. Complete gene annotation file (Ensembl GTF format) was extracted in order to determine sequence similarities and identify potential target sites in the coding regions. A BLASTN for FAD2-2 gene sequence has been established against Physcomitrella patens (Figure 1), a model plant which is widely known for targeted gene knockouts and facilitating reverse genetics approaches to study the physiology and development of plants. The indication of percentage identity was recorded as 99.2 % in the exonic regions.
Figure 1: The illustration of the result of Glycine max genome sequence homology against Physcomitrella patens using BLASTN type query. The genomic location was observed as 3 38046148 to 38047358(+). The recorded alignment score were 1171 with 99.2 similarities. Red color of the sequence indicated exons and blue indicates location of other alignments while other color represent markup loaded sequence.
The program SMS (Sequence Manipulation Suite) which is standardized by the ECMA (European Computer Manufacturers Association) reads the input target sequences and filter the target sequence by removing non-DNA characters from sequence to make it suitable for other potent applications. Sequence Manipulation Suite has been widely used for other significant conversion of different formats and sequence analysis including EMBL Trans Extractor, Shuffle DNA, Translation Map, Protein Isoelectric Point, and Pairwise Align DNA etc.
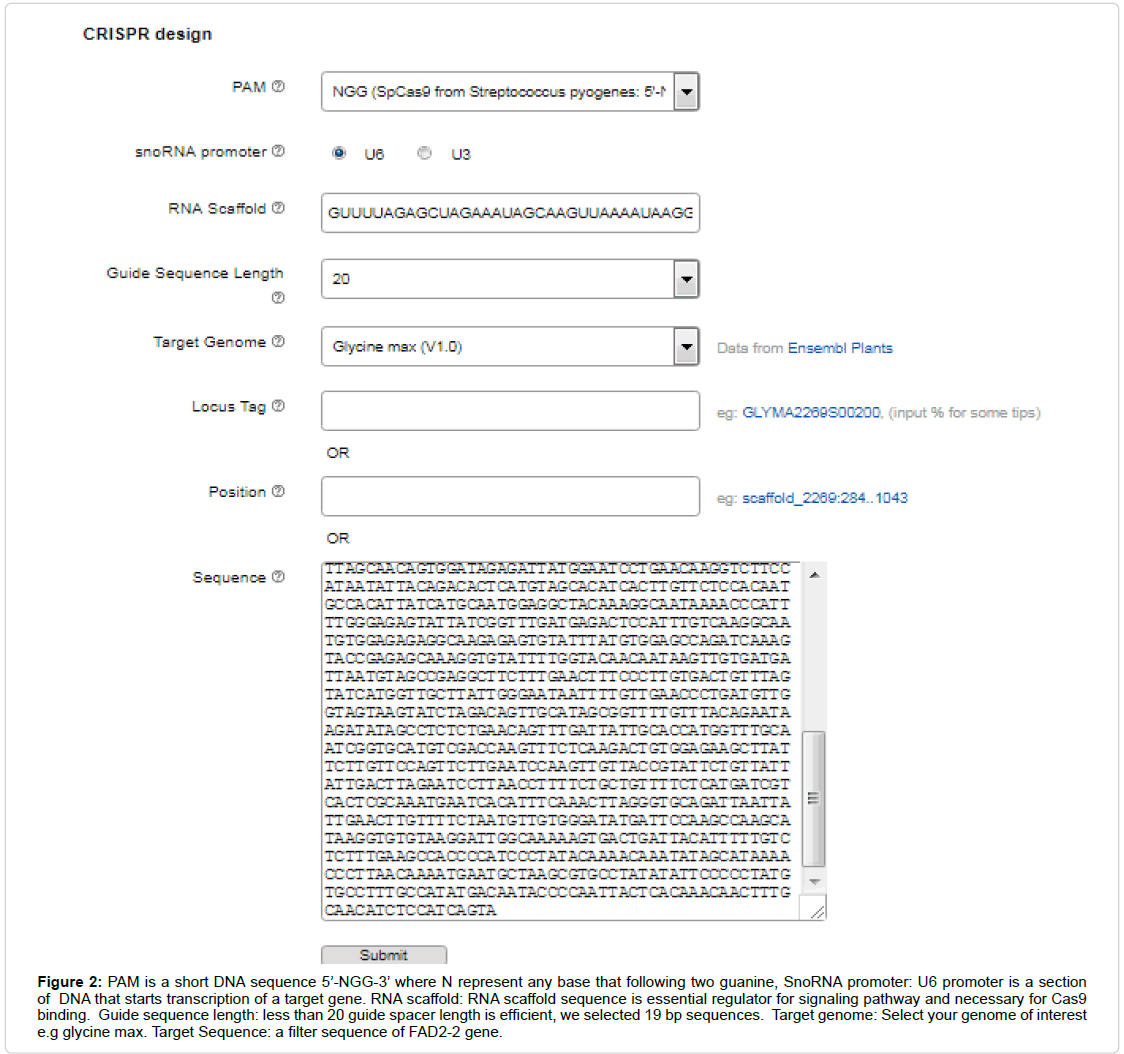
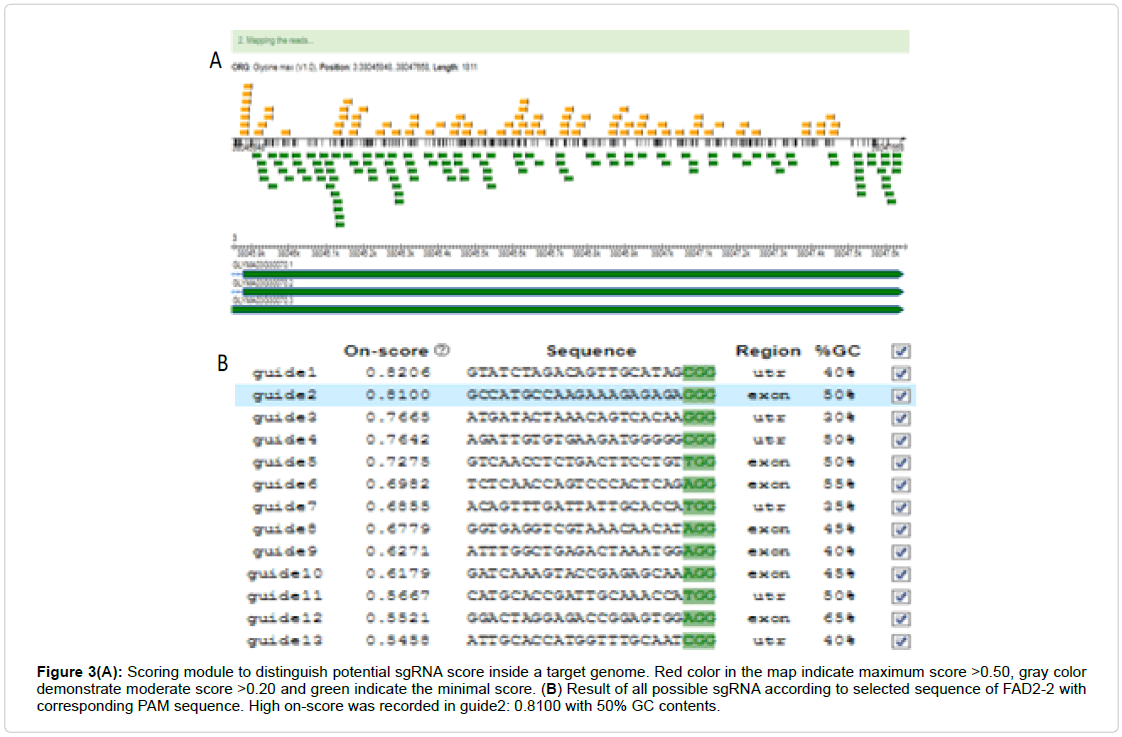
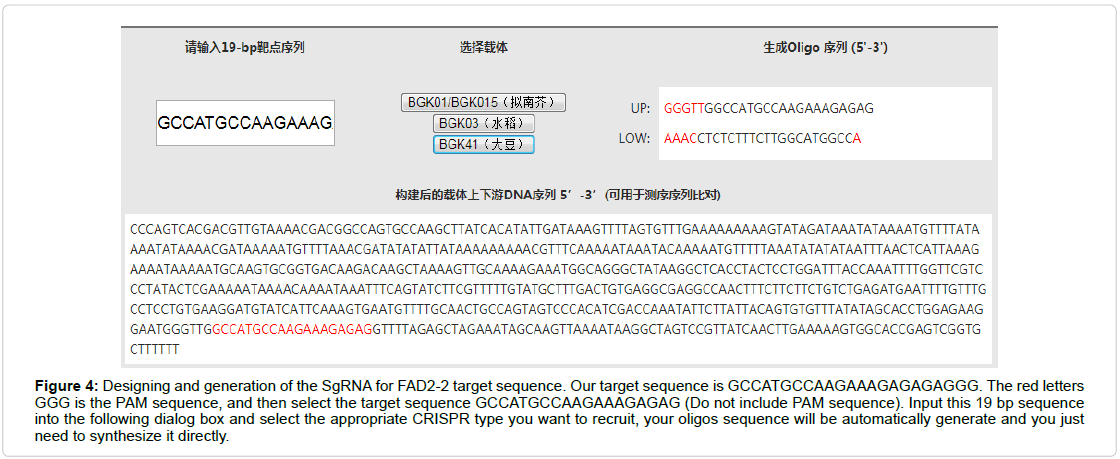
The CRISPR system required a routine SgRNA that contains a targeting sequence. The targeting sequence is homologous to your target gene or genomic region of interest. Online tool CRISPR P2 design was selected for SgRNA assembly for FAD2-2 sequence. Select the appropriate filter sequence in the range of 30-5,000 bps, select your genome of interest e.g. glycine max, other parameters such as PAM, snoRNA promoter, RNA scaffold, guide sequence (spacer) length are also require to be distinctly optimized (Figure 2). Then Submit the query and sequence map will be generated according to given genome (Figure 3A). After complete screening all possible SgRNA sequences are screen out with along with on target, as well as off target results including GC content and endonuclease sites (Figure 3B). Select 19 bp guide sequence indicating maximum score out of all possible sequences within exonic region (excluding PAM sequence). Customizing the target sequence of the SgRNA with the CRISPR vector construction kit Site involves designing a pair of oligos that correspond to the target genomic sequence (Figure 4).
Figure 2: PAM is a short DNA sequence 5’-NGG-3’ where N represent any base that following two guanine, SnoRNA promoter: U6 promoter is a section of DNA that starts transcription of a target gene. RNA scaffold: RNA scaffold sequence is essential regulator for signaling pathway and necessary for Cas9 binding. Guide sequence length: less than 20 guide spacer length is efficient, we selected 19 bp sequences. Target genome: Select your genome of interest e.g glycine max. Target Sequence: a filter sequence of FAD2-2 gene.
Figure 3(A): Scoring module to distinguish potential sgRNA score inside a target genome. Red color in the map indicate maximum score >0.50, gray color demonstrate moderate score >0.20 and green indicate the minimal score. (B) Result of all possible sgRNA according to selected sequence of FAD2-2 with corresponding PAM sequence. High on-score was recorded in guide2: 0.8100 with 50% GC contents.
Figure 4: Designing and generation of the SgRNA for FAD2-2 target sequence. Our target sequence is GCCATGCCAAGAAAGAGAGAGGG. The red letters GGG is the PAM sequence, and then select the target sequence GCCATGCCAAGAAAGAGAG (Do not include PAM sequence). Input this 19 bp sequence into the following dialog box and select the appropriate CRISPR type you want to recruit, your oligos sequence will be automatically generate and you just need to synthesize it directly.
Results and Customization
The complete cds of Glycine max FAD2-2 microsomal omega-6 desaturase mRNA in FASTA format has been shown here (Figure 1) which is extracted from GenBank (L43921.1). This 1501 bp long fragment is then subjected to BLASTN against Physcomitrella patens genome using ESNMBL PLANTS. The sequence similarities were recorded 99.1 % in the exonic regions chromosome number V1.0:3:38045848:38047658:1 loci.
The standardized ECMA (European Computer Manufacturers Association) program Sequence Manipulation Suite (SMS) was optimized for filtering non-DNA symbols like dots and numbering from aligned sequence. Filtering with SMS is mandatory before synthesizing oligos for sgRNA because it can influence the sequence of final output of oligos for targeted results. To accomplish the potential sites for sgRNA within FAD2-2 sequence the filtered sequence is further subjected to CRISPR-P, before jumping to CRISPR P 2.0 readers must care about certain parameters which is extremely sensitive and can influence the route of our sgRNA.
Designing of the optimal sgRNA in a filtered genome
Optimized CRISPR-P 2.0 has been designed for a total of 49 plant genomes which are previously sequenced. Step 1 Read the following guide steps to avoid biasness and to make sure the best possible results. Input the BLASTN sequence of target genome into Optimized CRISPR-P 2.0 software and select the target genome in our case study (Glycine max L.) for which we are intending to design possible SgRNA sequence (Figure 2) for more specific designing we can choose to provide either of these information like locus of our desired gene, the position of targeted chromosome or the complete preset nucleotide sequence of the gene. Apart from this some other parameters need to be optimized critically. Step 2: After submitting the work in a few seconds the user can get the SgRNA design page and the map of the projected genome (Figure 3A). And all the available SgRNA are showed on screen including on target score, GC content etc. We select a 19 bp high targeted SgRNA in the exonic region excluding PAM sequence for oligos designing (Figure 3B).
OLIGOS designing for FAD2-2 gene and cloning into CRISPR/ Cas BGK041 vector
After tailoring the possible target sequencing of sgRNA for FAD2-2 microsomal omega-6 desaturase mRNA which was supposed to be cloned in (NAME) CRISPR/Cas9 Vector it requires a specific pair of oligos that will interplay with the target genomic sequence. While synthesizing these oligos, we selected the CRISPR vector construction kit and resubmitted the guide sequence in CRISPR/Cas BGK041 Vector for the principle of oligos construction (Figure 4). The forward oligo (Oligo up) is composed of 19-nucleotide sequence (5’-GCCATGCCAAGAAAGAGAG-3’), immediately linked toward AGG Proto-spacer Adjacent Motif (PAM) sequence at the 3’ end of the desired sequence (excluding the PAM sequence). The reverse Oligo (Oligo Low) (5’-CTCTCTTTCTTGGCATGGC-3’) is the complementary of forward Oligo (Oligo up). To reduce the off target cleavage, the whole desirable sequence and PAM sequence must have at least three mismatches among any other genomic sequence [15]. It is mainly useful if mismatches be in the PAM site or adjacent toward the PAM site.
In vitro OLIGOS preparation and cloning into CRISPR/ Cas9 (BGK041)
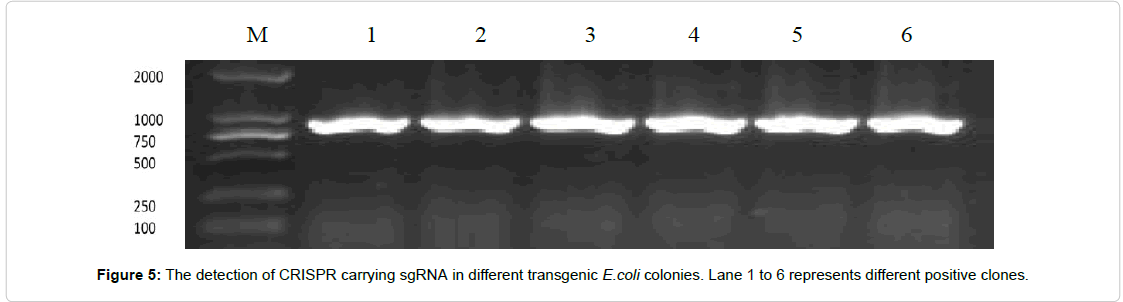
Synthesized targeted Oligos were dissolved in 10 μM nuclease free water followed by 3 minutes heat shock through PCR at 95ºC then slowly reduced the temperature up to 20ºC. The transformation of synthetic oligos into CRISPR Cas9 vector was performed according to manufacturer’s protocol with cold and heat shock method. Positive clones of E. coli carrying synthetic oligo in CRISPR Cas9 vector were detected on LB kanamycine resistant medium. Further detection was confirmed through Polymerase chain reaction using CRISPR Cas9 specific primers (Figure 5). The PCR reaction was followed with 35 cycles were followed as 94ºC for 30 s, 56ºC for 30s and 72ºC for 50s.
References
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, et al. (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816-821.
- Wiedenheft B, Sternberg SH, Doudna JA (2012) RNA-guided genetic silencing systems in bacteria and archaea. Nature482: 331-228.
- Cong L, Ran FA, Cox D, Lin S, Barretto R, et al. (2013) Multiplex genome engineering using CRISPR/Cas systems. Science339: 819-823.
- Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, et al. (2012) A programmable Double-RNA Guided DNA endonuclease in adaptive bacterial immunity. Science 337: 816-821.
- Puchta H, Fauser F (2014) Synthetic nucleases for genome engineering in plants: prospects for a bright future. Plant J 78: 727-741.
- Fan D, Liu T, Jiao B, Hou Y, Li S, et al. (2015) Efficient CRISPR/Cas9-mediated targeted Mutagenesis in Populus in the first generation. Sci Rep 5: 12217.
- Sun X, Hu Z, Chen R, Jiang Q, Song G, et al. (2015) Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci Rep 5: 10342.
- Cermak T, Baltes NJ, Cegan R, Zhang Y, Voytas DF (2015) High-frequency, precise modification of the tomato genome. Genome Biol 16: 232.
- Bortesi L, Fischer R (2015) The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol Adv 33: 41-52.
- Rinaldo AR, Ayliffe M (2015) Gene targeting and editing in crop plants: a new era of precision opportunities. Molecular Breeding 35: 40.
- Wang Y, Cheng X, Shan Q, Zhang Y, Liu J, et al. (2014) Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol 32: 947-951.
- Feng Z, Mao Y, Xu N, Zhang B, Wei P, et al. (2014) Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc Natl Acad Sci 111: 4632-4637.
- Zhang H, Zhang J (2014) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12: 797-807.
- Cradick TJ, Fine EJ (2013) CRISPR/Cas9 systems targeting beta-globin and CCR5genes have substantial off-target activity. Nucleic Acids Res 41: 9584-9592.
- Hsu PD, Scott DA (2013) DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol, 31: 827-832.