Research Article, J Genes Proteins Vol: 1 Issue: 1
DNA Barcoding of Freshwater Prawn Species of Two Genera Macrobrachium and Caridina Using mt-COI Gene
Udayasuriyan R, Saravana Bhavan P* and Kalpana R
Department of Zoology, Bharathiar University, Tamil Nadu, India
*Corresponding Author : Saravana Bhavan Periyakali
Department of Zoology, Bharathiar University, Coimbatore-641046, Tamil Nadu, India
Tel: +91-422-2428495
Fax: +91-422-2425706
E-mail: bhavan@buc.edu.in
Received: December 21, 2017 Accepted: December 23, 2017 Published: December 30, 2017
Citation: Udayasuriyan R, Saravana Bhavan P, Kalpana R (2017) DNA Barcoding of Freshwater Prawn Species of Two Genera Macrobrachium and Caridina Using mt-COI Gene. J Genes Proteins 1:1.
Abstract
Objective: The aim was to barcoding of Macrobrachium and Caridina species through mt-COI gene, and phylogenetic reconstruction based on the degree of genetic variability between species of these two genera.
Methods: Freshwater prawns were collected from few locations in the Cauvery River, Tamil Nadu, India. They were morphologically identified as Macrobrachium lamarrei, Macrobrachium lamarrei lamarroids, Macrobrachium malcolmsonii and Caridina gracilipes. Genomic DNA was isolated and amplification of mt-COI gene was done. The nucleotide divergence and some phylogenetic information were calculated by using MEGA v.6.01 and DAMBE. The phylognetic tree topology were reconstructed by Maximum likelihood model.
Results: The mt-COI gene sequences of these species showed 99-100% similarity. When Macrobrachium species were compared within themselves, they showed higher number of variable amino acid sites (539), which revealed some distance. When Macrobrachium species were compared with Caridina, they showed still higher number of variable amino acid sites (584), which revealed clear discrimination. In the subjected category, the mean value of inter-general divergence was more (6.934%) than that of the intra-general divergence (within Macrobrachium species, 3.628%; within M. lamarrei, 3.132%; within Caridina, 2.697%). When both subjected and retrieved species of these two genera were pooled together, the mean inter-general divergence value was also more (3.260%) than that of the intra-general divergence (Macrobrachium, 2.080%; within M. lamarrei, 1.222%; within Caridina, 2.200%). Therefore, the sequences of these two genera are more conserved as they showed 3.260-6.934% mean divergence or less subjected to evolutionary forces.
Conclusion: The species of Macrobrachium, and Caridina are clearly delineated from each other as the phylogenetic information obtained through mt-COI partial gene sequences are more conserved and less subjected to evolutionary forces, and thus, their species are genetically distinct.
Keywords: Mitochondrial COI Gene; Macrobrachium; Caridina; Divergence; Phylogenetic information; Tree topology
Introduction
The Cauvery River in India, contributes a considerable amount of both fin and shellfish in states of Tamil Nadu and Karnataka. Regarding diversity of Macrobrachium, seven species, such as Macrobrachium malcolmsonii, Macrobrachium nobilii, Macrobrachium scabriculum, Macrobrachium lamarrei, Macrobrachium rude, Macrobrachium australe and Macrobrachium aemulum have been reported in this river [1]. Among them M. malcolmsonii is the most widely distributed and holds first place in capture and culture fisheries. The presence of Caridina gracilipes, Macrobrachium malcolmsonii and Macrobrachium lamarrei in this river have been reported recently [2].
The phylogenetic affinities among Macrobrachium species are poorly understood. Pereira [3] carried out the first phylogenetic study based on morphological characters of the family Palaemonidae. Liu et al. [4] carried out molecular taxonomy of Macrobrachium. Baker et al. [5] highlighted the presence of several cryptic lineages in Australian Paratya (Atyidae), some of which represented as true species through DNA barcoding. Species discrimination has also been studied genetically in Caridina and cingeners from potential source populations throughout the west Indo-West Pacific region [6]. The nucleotide substitution is the main driving force for the formation of new species. The literature depict that the DNA sequence has been used to investigate the phylogenetic and biogeographic relationship among Macrobrachium canarae and Caridina gracilirostris [7]; the morphometric and meristic characters along with DNA bar-coding of CytB gene sequence has been used to discriminate Macrobrachium abrahami sp. [8]; the morphological, and partial sequence of mitochondrial COI gene (mt-COI) has been used to resolve the taxonomic identity of Caridina africana in the Cape Floristic Region of South Africa [9]; the mt-COI gene has also been used for phylogeography and population genetic studies of many freshwater shrimp species, due to the existence of high levels of sequence variability [10-12]; species discrimination in Penaeid shrimps, Macrobrachium, crabs and planktons through mt-COI gene have also been reported by us recently [2,13-17].
The time-series functions predict the prawn divergence. This is prevailing in organisms which undergo ontogenetic habitat shifts, and where the potentially limiting resources are age-specific, that is different life stages limited by different types of resources [18]. In some species, density dependence can particularly be strong during the early juvenile stage [19-21], whereas in others, later stages are more heavily influenced by density [20,22-25]. Such divergent patterns in the ontogenetic timing of density dependence may also exist among different populations within the same species [26,27].
Generally, the presence of plastic characters in the genus Macrobrachium makes the accurate determination of species more difficult and problematic, and the phylogenetics studies were poorly understood. These were shorted-out in this study while sequencing using COI gene, and have predicted some phylogenetic information led to well resolved tree topology. Actually, the present study describes the degree of genetic variability between Caridina and Macrobrachium species through mt-COI gene. Furthermore, the sequence similarity, amino acid residues, sequence divergence and phylogenetic information, such as synonymous and non-synonymous substitutions, transitional and transvertional substitutions, saturations and phylogenetic tree topology have also been studied.
Materials and Methods
Freshwater prawn species were collected from Hogenakkal (12.11° N, 77.77° E), Kooduthurai (11.09° N, 78.05° E), Aliyar Dam (10.48 N, 76.96 E) and Karungulam (10.35° E, 79.40° N). They were identified based on morphological characters, such as overlapping of the second segment over first and third segments, rostral structure, rostral teeth, periopods and telson with the help of experts, Mr. M. Kathirvel, Former Principal Scientist, Central Institute of Brackish water Aquaculture, ICAR, Chennai, India, and Mrs. K. Valarmathi, Scientist C, ZSI, Chennai, India. The species collected from Hogenakkal were morphologically identified as Macrobrachium lamarrei and Macrobrachium lamarrei lamarroids. The species collected from Kooduthurai were morphologically discriminated as Macrobrachium lamarrei and Caridina gracilipes. The species collected from Aliyar Dam were also morphologically discriminated as Macrobrachium lamarrei and Caridina gracilipes. Whereas, the species collected from Karungulum was morphologically identified as Macrobrachium malcolmsonii.
Genomic DNA was isolated from the abdominal tissue by using Qiagen DNeasy Blood and Tissue Kit (Germany). 1% Agarose Gel Electrophoresis were performed and the genomic DNA was detected in a Gel documentation system. DNA amplification of mt-COI gene was carried out with universal primers of forward and reverse in natures, COIa and COIf respectively. These primer sets were worked well with freshwater prawns [2]. The amplified product was resolved with 2% AGE, and they were sequenced. The forward and reverse sequences were aligned pairwise by using CAP3. The sequence similarity available with NCBI database was identified by BLAST. The internal stop codon was removed by using BLAST. The reading frame shift was detected by ORF finder. The trimmed sequence was authenticated with GenBank.
The multiple sequence alignment was done by using T-Coffee and the aligned sequence were highlighted with multiple align show (MAS) as identical, similar and variable sites of amino acids. The nucleotide composition (AT and GC bias), nucleotide divergence (K2P model) [28] and some phylogenetic information were calculated by using MEGA v.6.01. Assessment of synonymous (Ks) and nonsynonymous (Ka) substitutions for 3rd codon positions was calculated by Li93 method of DAMBE [29]. Similarly, the synonymous (Ka) and non-synonymous (Ks) substitutions for 3rd codon positions was predicted by Muse-Gaut model of codon substitution [30]. The transitional (Ts) and transvertional (Tv) substitutions was determined by the Felsenstein model of nucleotide substitution [31]. Analysis of sequence saturation, index of substitutional saturation (Iss) and critical value of index of substitutional saturation (Iss.c) was done by Xia method using DAMBE [32,33]. Finally the phylogenetic tree was reconstructed by Maximum Likelihood model [34,35].
Results and Discussion
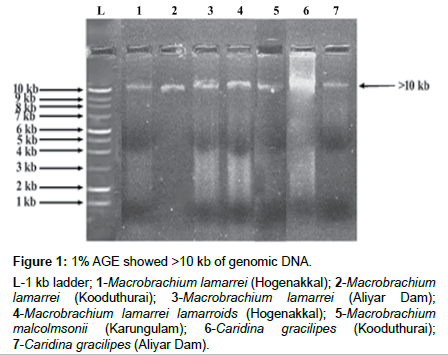
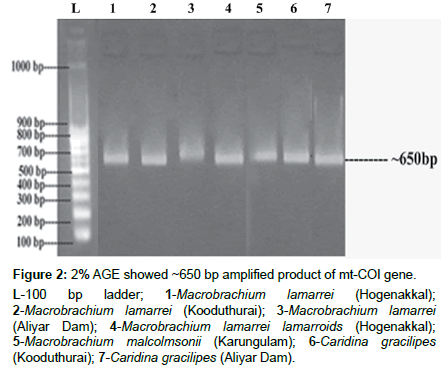
The isolated genomic DNA was confirmed as >10 kb and its amplified products showed ~650 bp (Figures 1 and 2). The actual length of the trimmed sequences were 443, 587, 657, 616, 598, 654 and 657 bp, for M. lamarrei (Hogenakkal), M. lamarrei (Kooduthurai), M. lamarrei (Aliyar Dam), M. lamarrei lamarroids (Hogenakkal), M. malcolmsonii (Karungulam), C. gracilipes (Kooduthurai) and C. gracilipes (Aliyar Dam) respectively, which were authenticated with GenBank (KX214617, KX788818, KX214618, KX788820, MF838938, KX788819 and KX214619 respectively).
Figure 1: 1% AGE showed >10 kb of genomic DNA.
L-1 kb ladder; 1-Macrobrachium lamarrei (Hogenakkal); 2-Macrobrachium lamarrei (Kooduthurai); 3-Macrobrachium lamarrei (Aliyar Dam); 4-Macrobrachium lamarrei lamarroids (Hogenakkal); 5-Macrobrachium malcolmsonii (Karungulam); 6-Caridina gracilipes (Kooduthurai); 7-Caridina gracilipes (Aliyar Dam).
Figure 2: 2% AGE showed ~650 bp amplified product of mt-COI gene.
L-100 bp ladder; 1-Macrobrachium lamarrei (Hogenakkal); 2-Macrobrachium lamarrei (Kooduthurai); 3-Macrobrachium lamarrei (Aliyar Dam); 4-Macrobrachium lamarrei lamarroids (Hogenakkal); 5-Macrobrachium malcolmsonii (Karungulam); 6-Caridina gracilipes (Kooduthurai); 7-Caridina gracilipes (Aliyar Dam).
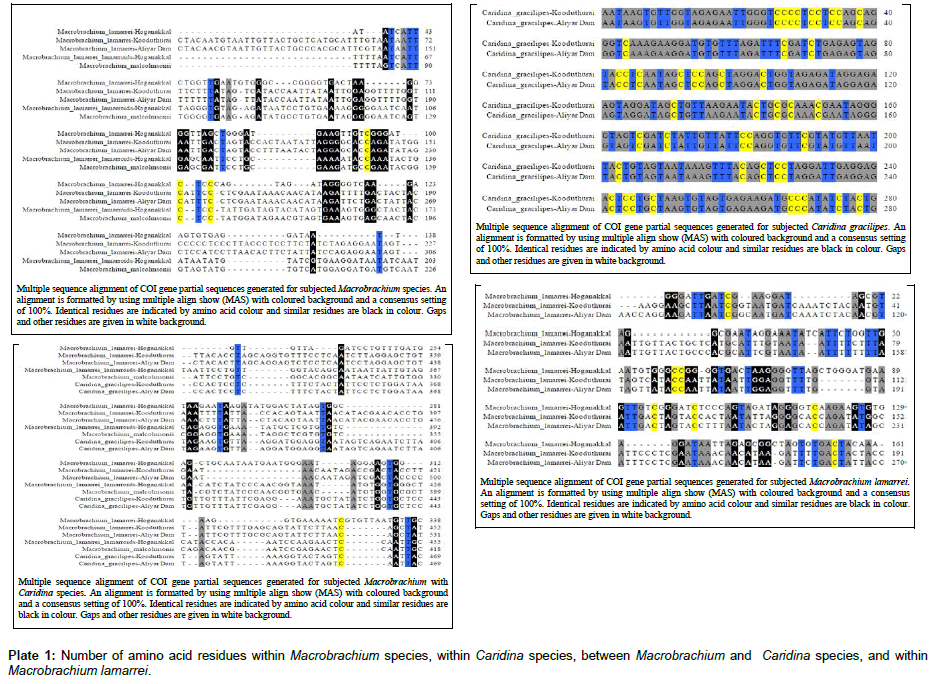
The BLAST results for these sequences showed 99-100% similarity with the retrieved sequences from the NCBI database. The similarity between the sequences usually depend entirely on the similarity in nucleotide frequencies, which is based on level of substitutional saturation, which in turn decreases phylogenetic information [32]. The results of multiple sequence alignment revealed the following, when Macrobrachium species were compared within themselves showed more number of variable amino acid sites (539). However, when Macrobrachium species were compared with Caridina, they showed still higher number of variable amino acid sites (584). Instead, Caridina species taken from two different locations showed more number of identical amino acid residues (654) and very less numbers of variable amino acid (4). Therefore, Caridina species showed a very close relationship within themselves, which indicates the fact that there was no sequential variations appeared even though they were taken from two different locations. Whereas, Macrobrachium species showed some distance within themselves because of more number of variable amino acids. Moreover, M. lamarrei taken from three locations also showed a higher number of variable amino acids (390) than that of identical amino acid (202), which indicates the fact that sequential variations occurred in the same species due to locality variation (Plate 1). At the outset, when the species of two genera, Macrobrachium and Caridina were compared, they showed much more variation in the amino acid sites, which indicate the fact that they are very well discriminated.
Nucleotide base composition
The base composition of the COI gene fragment varied among the species, AT biases was ranged from 55.4% to 62.7% (M. malcolmsonii and M. lamarrei lamarroids respectively) and the GC bias ranged between 37.3-44.6% (M. lamarrei lamarroids and M. malcolmsonii). The Macrobrachium species were compared within themselves showed 59.4% (A=30.4; T=29.0) and 40.6% of GC (G=22.0; C=18.6). The Caridina species were compared within themselves showed 59.8% AT bias (A=30; T=29.8) and 40.2% of GC (G=23; C=17.2). When Macrobrachium and Caridina were compared, they showed 59.5% AT bias (A=30.3; T=29.2) and 40.5% of GC (G=22.1; C=18.4). M. lamarrei within themselves showed 59.6% AT bias (A=30; T=29.5) and 40.3% of GC (G=20.5; C=19.8). In these four categories, collectively, the AT biases (60%) were more than that of the GC biases (40%). Therefore, no differences were seen in AT and GC biases between both genera. The higher AT bias recorded indicates the lower abundance of nuclear copies of mt-DNA (NUMTs) genes known as pseudogenes, homologs or paralogs in both genera (Table 1). The higher AT biases have also been reported by us in marine crabs [14], freshwater crabs and prawns [2,15] and freshwater plankton [16,17].
| Species name | Nucleotide % | |||||
|---|---|---|---|---|---|---|
| A | T | AT | G | C | GC | |
| Macrobrachium lamarrei of Hogenakkal | 30.9 | 28.7 | 59.6 | 27.5 | 12.9 | 40.4 |
| Macrobrachium lamarrei of Kooduthurai | 29.3 | 30.0 | 59.3 | 17.2 | 23.5 | 40.7 |
| Macrobrachium lamarrei of Aliyar Dam | 30.0 | 30.0 | 60.0 | 17.0 | 23.0 | 40.0 |
| Macrobrachium lamarrei lamarroids of Hogenakkal | 33.8 | 28.9 | 62.7 | 21.4 | 15.9 | 37.3 |
| Macrobrachium malcolmsonii of Karungulam | 27.9 | 27.4 | 55.4 | 26.9 | 17.7 | 44.6 |
| Caridina gracilipes of Kooduthurai | 30.1 | 29.7 | 59.8 | 22.9 | 17.3 | 40.2 |
| Caridina gracilipes of Aliyar Dam | 30.0 | 29.8 | 59.8 | 23.0 | 17.2 | 40.2 |
| Within Macrobrachium species | 30.4 | 29.0 | 59.4 | 22.0 | 18.6 | 40.6 |
| Within Caridina gracilipes | 30.0 | 29.8 | 59.8 | 23.0 | 17.2 | 40.2 |
| Between Macrobrachium and Caridina species | 30.3 | 29.2 | 59.5 | 22.1 | 18.4 | 40.5 |
| Within M. lamarrei | 30.0 | 29.5 | 59.6 | 20.5 | 19.8 | 40.3 |
Table 1: Nucleotide base composition percentage in COI partial gene sequences of subjected Macrobrachium and Caridina species.
Nucleotide divergence
The nucleotide divergence between subjected species, and between subjected and retrieved species are depicted in Table 2. In the subjected category, the mean divergent rate of different Macrobrachium species was 3.628 witha maximum of 15.363 (between M. malcolmsonii and M. lamarrei) and minimum of 0.701 (between M. lamarrei and M. lamarrei lamarroids). The divergence of Caridina species within themselves was 2.697. When Macrobrachium and Caridina species were combined in one category, the mean divergence value was 6.934 with a maximum of 23.268 and minimum of 1.026. M. lamarrei alone showed 3.132% of mean divergence value within themselves taken from three different locations with a maximum of 5.370 and minimum of 1.707. Therefore, between two genera, Macrobrachium and Caridina the divergence was >3.0% in most of the cases. However, in few cases the divergence value was <3.0%, where the morphological characters play a vital role in species discrimination. In contrast to this, when the same species of different locality showed >3.0% divergence value (M. lamarrei of Kooduthurai vs. M. lamarrei of Aliyar Dam) they may be either sub-species of cryptic in nature. When retrieved species are included with subjected species >3% divergence value appeared at 12 combinations, of which 4 in within Macrobrachium species and 8 in Macrobrachium and Caridina, and none of the combinations within Caridina as well as within M. lamarrei showed >3.0% variation. The interspecific distance ranged from 13.2-19.9% between the species, Macrobrachium borellii, Macrobrachium brasiliense, Macrobrachium jelskii, Macrobrachium olfersii, Macrobrachium crenulatum, Macrobrachium americanum, Macrobrachium carcinus and Macrobrachium acanthurus, and the intraspecific distance ranged from 0-3.3% among different population of Macrobrachium amazonicum has been reported using COI sequences [36].
| Comparisons | Divergence (%) |
|---|---|
| Within Macrobrachium species (subjected) | |
| Macrobrachium lamarrei of Hogenakkal vs. Macrobrachium lamarrei of Kooduthurai | 2.321 |
| Macrobrachium lamarrei of Hogenakkal vs. Macrobrachium lamarrei of Aliyar Dam | 1.707 |
| Macrobrachium lamarrei of Hogenakkal vs. Macrobrachium lamarrei lamarroids of Hogenakkal | 1.182 |
| Macrobrachium lamarrei of Hogenakkal vs. Macrobrachium malcolmsonii of Karungulam | 2.583 |
| Macrobrachium lamarrei of Kooduthurai vs. Macrobrachium lamarrei lamarroids of Hogenakkal | 1.182 |
| Macrobrachium lamarrei of Kooduthurai vs. Macrobrachium lamarrei of Aliyar Dam | 5.370 |
| Macrobrachium lamarrei of Kooduthurai vs. Macrobrachium malcolmsonii of Karungulam | 2.583 |
| Macrobrachium lamarrei lamarroids of Hoganakkal vs. Macrobrachium lamarrei of Aliyar Dam | 0.701 |
| Macrobrachium malcolmsonii of Karungulam vs. Macrobrachium lamarrei of Aliyar Dam | 15.363 |
| Macrobrachium lamarrei lamarroids of Hogenakkal vs. Macrobrachium malcolmsonii of Karungulam | 3.293 |
| Mean Divergence | 3.628 |
| Within Caridina species (subjected) | |
| Caridina gracilipes of Kooduthurai vs. Caridina gracilipes of Aliyar Dam | 2.697 |
| Between Macrobrachium and Caridina species (subjected) | |
| Macrobrachium lamarrei of Hogenakkal vs. Caridina gracilipes of Kooduthurai | 1.026 |
| Macrobrachium lamarrei of Kooduthurai vs. Caridina gracilipes of Kooduthurai | 5.990 |
| Macrobrachium lamarrei of Aliyar Damvs. Caridina gracilipes of Kooduthurai | 23.268 |
| Macrobrachium lamarrei lamarroids of Hogenakkal vs. Caridina gracilipes of Kooduthurai | 3.074 |
| Macrobrachium malcolmsonii of Karungulam vs. Caridina gracilipes of Kooduthurai | 1.068 |
| Macrobrachium lamarrei of Hogenakkal vs. Caridina gracilipes of Aliyar Dam | 1.026 |
| Macrobrachium lamarrei of Kooduthurai vs. Caridina gracilipes of Aliyar Dam | 5.990 |
| Macrobrachium lamarrei of Aliyar Dam vs. Caridina gracilipes of Aliyar Dam | 23.268 |
| Macrobrachium lamarrei lamarroidsof Hogenakkal vs. Caridina gracilipes of Aliyar Dam | 3.564 |
| Macrobrachium malcolmsonii of Karungulam vs. Caridina gracilipes of Aliyar Dam | 1.068 |
| Mean Divergence | 6.934 |
| Within M. lamerrei (subjected) | |
| Macrobrachium lamarrei of Hogenakkal vs. Macrobrachium lamarrei of Kooduthurai | 2.321 |
| Macrobrachium lamarrei of Hogenakkal vs. Macrobrachium lamarrei of Aliyar Dam | 1.707 |
| Macrobrachium lamarrei of Kooduthurai vs. Macrobrachium lamarrei of Aliyar Dam | 5.370 |
| Mean Divergence | 3.132 |
| Within Macrobrachium species (subjected and retrieved) | |
| Macrobrachium lamarrei of Hogenakkal vs. Macrobrachium lamarrei (India) | 0.291 |
| Macrobrachium lamarrei of Kooduthurai vs. Macrobrachium lamarrei (India) | 0.678 |
| Macrobrachium lamarrei of Aliyar Dam vs. Macrobrachium lamarrei (India) | 2.697 |
| Macrobrachium lamarrei lamarroids of Hogenakkal vs. Macrobrachium lamarrei (India) | 2.813 |
| Macrobrachium malcolmsonii of Karungulam vs. Macrobrachium lamarrei (India) | 2.665 |
| Macrobrachium lamarrei of Hogenakkal vs. Macrobrachium malcolmsonii (Tamil Nadu, India) | 0.288 |
| Macrobrachium lamarrei of Kooduthurai vs. Macrobrachium malcolmsonii (Karala, India) | 2.813 |
| Macrobrachium lamarrei of Aliyar Dam vs. Macrobrachium malcolmsonii (Tamil Nadu, India) | 2.665 |
| Macrobrachium lamarrei lamarroids of Hogenakkal vs. Macrobrachium malcolmsonii (Karala, India) | 3.583 |
| Macrobrachium malcolmsonii of Karungulam vs. Macrobrachium malcolmsonii (India) | 2.697 |
| Macrobrachium lamarrei of Hogenakkal vs. Macrobrachium malcolmsonii (Karala, India) | 0.128 |
| Macrobrachium lamarrei of Kooduthurai vs. Macrobrachium malcolmsonii (Tamil Nadu, India) | 3.023 |
| Macrobrachium lamarrei of Aliyar Dam vs. Macrobrachium malcolmsonii (Kerala, India) | 4.200 |
| Macrobrachium lamarrei lamarroids of Hogenakkal vs. Macrobrachium malcolmsonii (India) | 0.620 |
| Macrobrachium malcolmsonii of Karungulam vs. Macrobrachium malcolmsonii (India) | 0.706 |
| Macrobrachium malcolmsonii (Tamil Nadu, India) vs. Macrobrachium lamarrei (India) | 2.665 |
| Macrobrachium malcolmsonii (Kerala, India) vs. Macrobrachium lamarrei (India) | 4.200 |
| Macrobrachium malcolmsonii (India) vs. Macrobrachium malcolmsonii (India) | 0.706 |
| Mean Divergence | 2.080 |
| Within Caridina species (subjected and retrieved) | |
| Caridina gracilipes of Kooduthurai vs. Caridina gracilipes (India) | 2.260 |
| Caridina gracilipes of Aliyar Dam vs. Caridina gracilipes (China) | 2.260 |
| Caridina gracilipes of Kooduthurai vs. Caridina gracilipes (India) | 2.126 |
| Caridina gracilipes of Aliyar Dam vs. Caridina gracilipes (China) | 2.126 |
| Caridina gracilipes (India) vs. Caridina gracilipes (China) | 1.733 |
| Mean Divergence | 2.200 |
| Macrobrachiumand Caridina species (subjected and retrieved) | |
| Macrobrachium lamarrei of Hogenakkal vs. Macrobrachium lamarrei of Kooduthurai | 2.321 |
| Macrobrachium lamarrei of Hogenakkal vs. Macrobrachium lamarrei of Aliyar Dam | 1.707 |
| Macrobrachium lamarrei of Hogenakkal vs. Macrobrachium lamarrei lamarroids of Hogenakkal | 1.182 |
| Macrobrachium lamarrei of Hogenakkal vs. Macrobrachium malcolmsonii of Karungulam | 2.583 |
| Macrobrachium lamarrei of Kooduthurai vs. Macrobrachium lamarrei lamarroids of Hogenakkal | 1.182 |
| Macrobrachium lamarrei of Kooduthurai vs. Macrobrachium lamarrei of Aliyar Dam | 5.37 |
| Macrobrachium lamarrei of Kooduthurai vs. Macrobrachium malcolmsonii of Karungulam | 2.583 |
| Macrobrachium lamarrei of Aliyar Damvs. Macrobrachium lamarrei lamarroids of Hogenakkal | 0.701 |
| Macrobrachium lamarrei of Aliyar Dam vs. Macrobrachium malcolmsonii of Karungulam | 15.363 |
| Macrobrachium lamarrei lamarroids of Hogenakkal vs. Macrobrachium malcolmsonii of Karungulam | 3.293 |
| Caridina gracilipes of Kooduthurai vs. Caridina gracilipes of Aliyar Dam | 2.697 |
| Macrobrachium lamarrei of Hogenakkal vs. Caridina gracilipes of Kooduthurai | 1.026 |
| Macrobrachium lamarrei of Kooduthurai vs. Caridina gracilipes of Kooduthurai | 5.99 |
| Macrobrachium lamarrei of Aliyar Damvs. Caridina gracilipes of Kooduthurai | 23.268 |
| Macrobrachium lamarrei lamarroids of Hogenakkal vs. Caridina gracilipes of Kooduthurai | 3.074 |
| Macrobrachium malcolmsonii of Karungulam vs. Caridina gracilipes of Kooduthurai | 1.068 |
| Macrobrachium lamarrei of Hogenakkal vs. Caridina gracilipes of Aliyar Dam | 1.026 |
| Macrobrachium lamarrei of Kooduthurai vs. Caridina gracilipes of Aliyar Dam | 5.99 |
| Macrobrachium lamarrei of Aliyar Dam vs. Caridina gracilipes of Aliyar Dam | 23.268 |
| Macrobrachium lamarrei lamarroids of Hogenakkal vs. Caridina gracilipes of Aliyar Dam | 3.564 |
| Macrobrachium malcolmsonii of Karungulam vs. Caridina gracilipes of Aliyar Dam | 1.068 |
| Macrobrachium lamarrei of Hogenakkal vs. Macrobrachium lamarrei (India) | 0.291 |
| Macrobrachium lamarrei of Kooduthurai vs. Macrobrachium lamarrei (India) | 0.678 |
| Macrobrachium lamarrei of Aliyar Dam vs. Macrobrachium lamarrei (India) | 2.697 |
| Macrobrachium lamarrei lamarroids of Hogenakkal vs. Macrobrachium lamarrei (India) | 2.813 |
| Macrobrachium malcolmsonii of Karungulam vs. Macrobrachium lamarrei (India) | 2.665 |
| Macrobrachium lamarrei of Hogenakkal vs. Macrobrachium malcolmsonii (Tamil Nadu, India) | 0.288 |
| Macrobrachium lamarrei of Kooduthurai vs. Macrobrachium malcolmsonii (Kerala, India) | 2.813 |
| Macrobrachium lamarrei of Aliyar Dam vs. Macrobrachium malcolmsonii (Tamil Nadu, India) | 2.665 |
| Macrobrachium lamarrei lamarroids of Hogenakkal vs. Macrobrachium malcolmsonii (Kerala, India) | 3.583 |
| Macrobrachium malcolmsonii of Karungulam vs. Macrobrachium malcolmsonii (Tamil Nadu, India) | 2.697 |
| Macrobrachium lamarrei of Hogenakkal vs. Macrobrachium malcolmsonii (Kerala, India) | 0.128 |
| Macrobrachium lamarrei of Kooduthurai vs. Macrobrachium malcolmsonii (Tamil Nadu, India) | 3.023 |
| Macrobrachium lamarrei of Aliyar Dam vs. Macrobrachium malcolmsonii (Kerala, India) | 4.200 |
| Macrobrachium lamarrei lamarroids of Hogenakkal vs. Macrobrachium malcolmsonii (Tamil Nadu, India) | 0.620 |
| Macrobrachium malcolmsonii of Karungulam vs. Macrobrachium malcolmsonii (Kerala, India) | 0.706 |
| Caridina gracilipes of Kooduthurai vs. Caridina gracilipes (India) | 2.26 |
| Caridina gracilipes of Aliyar Dam vs. Caridina gracilipes (China) | 2.26 |
| Caridina gracilipes of Kooduthurai vs. Caridina gracilipes (India) | 2.126 |
| Caridina gracilipes of Aliyar Dam vs. Caridina gracilipes (China) | 2.126 |
| Macrobrachium lamarrei of Hogenakkal vs. Caridina gracilipes (India) | 0.024 |
| Macrobrachium lamarrei of Kooduthurai vs. Caridina gracilipes (China) | 3.561 |
| Macrobrachium lamarrei of Aliyar Dam vs. Caridina gracilipes (India) | 4.926 |
| Macrobrachium lamarrei lamarroids of Hogenakkal vs. Caridina gracilipes (India) | 0.823 |
| Macrobrachium malcolmsonii of Karungulam vs. Caridina gracilipes (India) | 0.945 |
| Macrobrachium lamarrei of Hogenakkal vs. Caridina gracilipes (China) | 0.500 |
| Macrobrachium lamarrei of Kooduthurai vs Caridina gracilipes (China) | 4.309 |
| Macrobrachium lamarrei of Aliyar Dam vs. Caridina gracilipes (China) | 3.460 |
| Macrobrachium lamarrei lamarroids of Hogenakkal vs. Caridina gracilipes (China) | 1.313 |
| Macrobrachium malcolmsonii of Karungulam vs. Caridina gracilipes (China) | 4.587 |
| Macrobrachium lamarrei (India) vs Caridina gracilipes of Kooduthurai | 1.987 |
| Macrobrachium lamarrei (India) vs. Caridina gracilipes of Aliyar Dam | 1.063 |
| Macrobrachium malcolmsonii ( India) vs. Caridina gracilipes of Kooduthurai | 1.572 |
| Macrobrachium malcolmsonii (India) vs. Caridina gracilipes of Aliyar Dam | 1.572 |
| Macrobrachium malcolmsonii (India) vs. Caridina gracilipes of Kooduthurai | 2.504 |
| Macrobrachium malcolmsonii (India) vs. Caridina gracilipes of Aliyar Dam | 2.504 |
| Mean Divergence | 3.260 |
| Within M. lamerrei (subjected and retrieved) | |
| Macrobrachium lamarrei of Hogenakkal vs. Macrobrachium lamarrei (India) | 0.291 |
| Macrobrachium lamarrei of Kooduthurai vs. Macrobrachium lamarrei (India) | 0.678 |
| Macrobrachium lamarrei of Aliyar Dam vs. Macrobrachium lamarrei (India) | 2.697 |
| Mean Divergence | 1.222 |
Table 2: Nucleotide divergence of Macrobrachium and Caridina species.
Phylogenetic information and tree topology
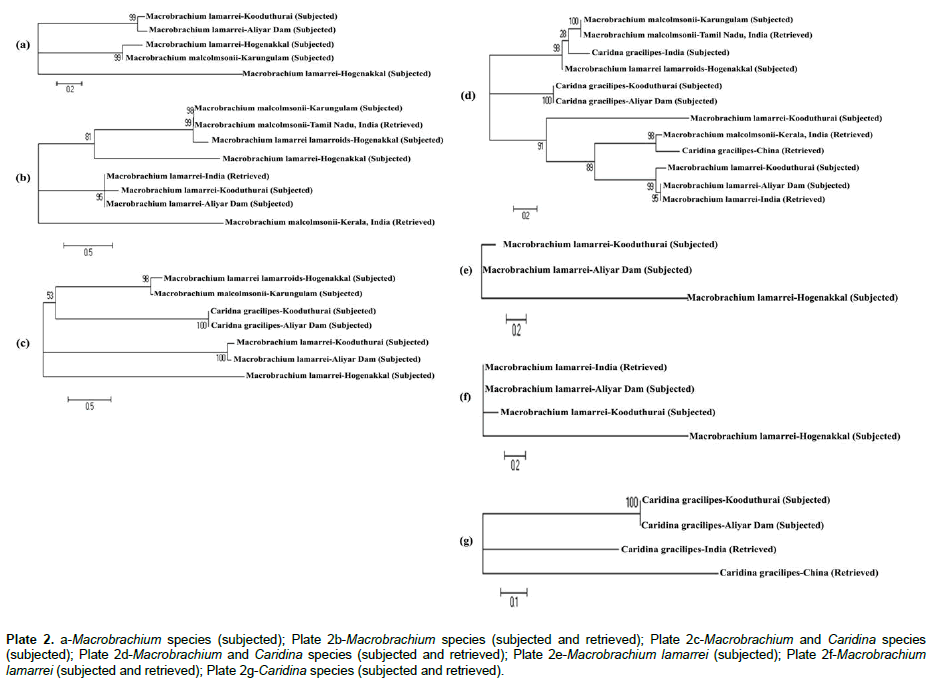
The predicted phylogenetic information, such as synonymous (Ks) and non-synonymous (Ka) substitutions, transitional (Ts) and transvertional (Tv) substitutions, and saturation, index of substitutional saturation (Iss) and critical value of index of substitutional saturation (Iss.c) are presented in Table 3. In the subjected category, when Macrobrachium species were compared within themselves, the Ka was higher (2.181) than that of Ks (0.739), which indicates the possibility of occurrence of more deleterious mutation and less silent mutation. Similarly, the Tv was higher (0.37) than that of Ts (0.26), which indicates the fact that these sequences have more phylogenetic information. Thus, little saturation occurred in these sequences, which was confirmed by the predicted lower Iss.c value (0.790) than that of the Iss (0.916) and therefore, little phylogenetic differences existed between sequences. When more species were included by retrieving, the sequences seemed to be saturated with more phylogenetic information, because the Iss.c was higher (0.935) than that of the Iss (0.777). The phylogenetic tree topology of Macrobrachium species (subjected) formed one clade, in which, M. lamarrei was aligned alone at the base of the tree and two clusters, of which one was formed by M. malcolmsonii and M. lamarrei lamarroids as sister taxa and the another was formed by M. lamarrei taken from two different places as sister taxon at the top of the tree (Plate 2a). When retrieved Macrobrachium was included with subjected Macrobrachium, a polyphyletic tree was appeared with two clades and two clusters. At the base, M. malcolmsonii (retrieved) was formed a clade, and the next clade with M. lamarrei (subjected) was formed in between two clusters. The first cluster consisted of M. lamarrei only, of which two subjected and one retrieved. The second cluster was formed by M. lamarrei lamarroids (subjected) and M. malcolmsonii (one subjected and one retrieved) (Plate 2b).
Plate 2: a-Macrobrachium species (subjected); Plate 2b-Macrobrachium species (subjected and retrieved); Plate 2c-Macrobrachium and Caridina species (subjected); Plate 2d-Macrobrachium and Caridina species (subjected and retrieved); Plate 2e-Macrobrachium lamarrei (subjected); Plate 2f-Macrobrachium lamarrei (subjected and retrieved); Plate 2g-Caridina species (subjected and retrieved).
| Phylogenetic information | Ks | Ka | Ks-Ka | Ts | Tv | Tv-Ts | Iss | Iss.c | Iss.c-Iss | |
|---|---|---|---|---|---|---|---|---|---|---|
| Subjected species alone |
Macrobrachium | 0.739 | 2.181 | 1.442 | 0.26 | 0.37 | 0.11 | 0.916 | 0.79 | 0.126 |
| Macrobrachium and Caridina | 0.719 | 2.178 | 1.459 | 0.27 | 0.40 | 0.13 | 0.768 | 0.886 | 0.118 | |
| M. lamarrei | 0.761 | 2.165 | 1.404 | 0.23 | 0.33 | 0.10 | 0.760 | 0.812 | 0.052 | |
| Subjected and retrieved species | Macrobrachium | 0.661 | 2.235 | 1.574 | 0.28 | 0.41 | 0.13 | 0.777 | 0.935 | 0.158 |
| Caridina | 0.665 | 2.238 | 1.573 | 0.24 | 0.36 | 0.12 | 0.702 | 0.806 | 0.104 | |
| Macrobrachium and Caridina | 0.693 | 2.210 | 1.517 | 0.29 | 0.43 | 0.14 | 0.756 | 1.062 | 0.306 | |
| M. lamarrei | 0.704 | 2.201 | 1.497 | 0.29 | 0.39 | 0.10 | 0.615 | 0.805 | 0.190 | |
Table 3: Overall average phylogenetic information of subjected and retrieved Macrobrachium and Caridina species.
When the species of two genera, Macrobrachium and Caridina were combined, the Ka was higher (2.178) than that of Ks (0.719), which also indicates the possibility of occurrence of more deleterious mutation than that of the silent mutation. Similarly, the Tv was also higher (0.40) than that of Ts (0.27), which indicates the fact that these sequences have still more phylogenetic information. However, saturation might have not been occurred in these sequences, because of two different genera, which was confirmed by the predicted higher Iss.c value (0.886) than that of the Iss (0.768) and more phylogenetic differences existed between sequences. When retrieved species of Macrobrachium and Caridina were included with the subjected species, the sequences showed no saturation with very highest Iss.c (1.062), which indicates more phylogenetic information in this study (Table 3). Therefore, both generic and species differences existed due to genetic differences, which in turn lead to species discrimination. The phylogenetic tree topology constructed with subjected species of Macrobrachium and Caridina formed one clade with M. lamarrei at the base and three clusters as sister taxon each with two species. The first cluster was formed by M. lamarrei, the second by C. gracilipes, and the third by M. malcolmsonii and M. lamarrei lamarroids at the top of the tree (Plate 2c). When both subjected and retrieved species of Macrobrachium and Caridina were taken in one group four clusters and a clade were formed. The first cluster was formed by M. lamarrei (one retrieved and two subjected). The second cluster was formed by two retrieved species (M. malcolmsonii and C. gracilipes) as sister taxon. Then a single distinct clade with subjected species of M. lamarrei was formed. The third cluster was formed by the subjected C. gracilipes. And the forth cluster was formed by M. lamarrei lamarroids (subjected), C. graclipes (retrieved) and M. malcolmsonii (both retrieved and subjected) as sister taxon (Plate 2d).
In the case of M. lamarrei taken from three different locations, the Ka was higher (2.165) than that of Ks (0.761), which also indicates the possibility of occurrence of more deleterious mutation in the sequences. Similarly, the Tv was also higher (0.33) than that of Ts (0.23), which indicates that these sequences have phylogenetic information. Though the Iss.c was higher (0.812) than that of the Iss (0.760), little saturation seemed to occur between these sequences, because of the same species, which showed only little difference between Iss.c and Iss (0.052) (Table 3). When retrieved species of M. lamarrei were included the sequences also showed little saturation with more phylogenetic information (Iss.c, 805; Iss, 615) (Table 3) due to locality/ country variation. The phylogenetic tree topology of subjected M. lamarrei formed a separate clade and a cluster (Plate 2e). When retrieved species of M. lamarrei was included, it joint in the cluster at the top (Plate 2f).
The phylogenetic information for subjected Caridina species was not predicted because of the less sampling, but, this was calculated when the retrieved Caridina species were included and they also showed little saturation with some phylogenetic information (Iss.c, 806; Iss, 702) (Table 3) due to locality/ country variation. The phylogenetic tree of C. gracilipes (both subjected and retrieved) formed two separate clades with retrieved species and a cluster of subjected species at the top (Plate 2g).
Species that have a wide distribution, in heterogeneous or geographically isolated environments can have a phenotype variation, because they are prone to show plastic responses to different environmental influences. The presence of plastic characters in the genus Macrobrachium makes the accurate determination of species more difficult and problematic [37,38]. The environmentdependent plasticity and the phenotypic variations stem from genetic or behavior differences between individuals are from ontogenetic developmental or combining of all these factors [39]. On the other hand, morphological characters may often be undergoing convergent evolution as they are under similar selective pressure [40]. The species of the genus Macrobrachium have high intraspecific variation and a single species may have genetic diversity and structured populations . A wide distribution and a great morphological variation during ontogenesis including the possibility of morphotypes within the species have been reported in congeneric species such as Macrobrachium rosenbergii [41], M. amazonicum [42], M. grandimanus [43-45] and Macrobrachium olfersii [46-49].
The development of an organism (ontogeny) expresses all the intermediate forms of its ancestors throughout evolution (phylogeny). Freshwater prawns of the genus Macrobrachium [50] (Crustacea: Palaemonidae) are a highly diverse group of decapod crustaceans thought to have originated from marine ancestors, some of which subsequently migrated towards fresh water in more than 1 wave; hence its members are known to inhabit the entire range of habitats from purely marine areas to inland hill streams and impounded water bodies [51-53]. In the present study, Macrobrachium showed > 3.0% interspecies divergence. It has been suggested that origin of Macrobrachium occurred in the late Oligocene or early Miocene [54]. It has been reported that species of palaemonoid, atyoid and alpheoid are not closely related [55]. Atyidae are the ancient inhabitants of freshwater, having diverged early from an ancestral marine stock [56]. Therefore, in this study, Macrobrachium and Caridina showed some distance. However, in Caridina, only one species, C. gracilipes was studied, thus, it is not possible to detect the plasticity. Moreover, it is important to mention here that very little is known on the phylogeny of Caridea due to the paucity of higher level cladistic and genetic studies [3].
Conclusion
In this study, the sequences of Macrobrachium and Caridina showed considerable degree of variation in the amino acid sites, which indicate that they were very well discriminated. The observed > 60% base composition indicates lower abundance of NUMTs genes. The >3.0% mean divergence recorded between different species of Macrobrachium indicates their clear discrimination. The subjected C. gracilipes always sat with a separate cluster and only the retrieved C. gracilipes was aligned with M. malcolmsonii and M. lamarrei lamarroids, and mostly M. lamarrei formed a separate cluster or clade. Therefore, the species of Macrobrachium, and Caridina are clearly delineated from each other as the phylogenetic information obtained through mt-COI partial gene sequences are more conserved and less subjected to evolutionary forces, and thus, these species are genetically distinct.
Acknowledgement
The Science and Engineering Research Board, Department of Science and Technology, Government of India, is gratefully acknowledged for the financial support provided in the form of research project (SB/EMEQ-291/2013 of the SERB, New Delhi).
References
- Mariappan P (2000) Studies on Chela biology and behaviour of Macrobrachium nobilii with special reference to Aquaculture. PhD dissertation, Bharathidasan University, Tiruchirapalli, India.
- Udayasuriyan R, Bhavan PS, Vadivalagan C, Rajkumar G (2015) Efficiency of different COI markers in DNA barcoding of freshwater prawn species. J Entomol Zool Stud 3: 98-110.
- Pereira G (1997) A cladistic analysis of the freshwater shrimps of the family Palaemonidae (Crustacea, Decapoda, Caridea). Acta Biol Venezulica 17: 1-69.
- Liu MY, Cai Y, Tzeng CS (2007) Molecular systematics of the freshwater prawn genus Macrobrachium Bate, 1868 (Crustacea: Decapoda: Palaemonidae) inferred from mtDNA sequences, with emphasis on East Asian species. Zool Stud46: 272-289.
- Baker AM, Hurwood DA, Krogh M, Hughes JM (2004) Mitochondrial DNA signatures of restricted gene flow within divergent lineages of an Atyid shrimp (Paratya australiensis). Heredity 93: 196-207.
- Page TJ, Choy SC, Hughes JM (2005) The taxonomic feedback loop: symbiosis of morphology and molecules. BiolLett 139-142.
- Siva Ranjanee S, Mariapan N (2011) A genetical and ecological diversity of fresh water prawns Macrobrachium canarae and Caridina gracilirostris from Kanyakumari Dist., Tamil Nadu, India. Int J Gen Eng Biotech 2: 23-32.
- Pillai PM, Unnikrishnan V, Suresh kumar U (2014) Description, DNA barcode and phylogeny of a new species, Macrobrachium abrahami (Decapoda: Palaemonidae) from Kerala, India. Zootaxa 3768: 546-556.
- Mirimin L, Kitchin N, Impson DN, Clark PF, Jasmine R, et al. (2015) Genetic and morphological characterization of freshwater shrimps (Caridina africanaKingsley, 1882) reveals the presence of alien shrimps in the Cape Floristic region, South Africa. J Heredity 711-718.
- Cook BD, Baker AM, Page TJ, Grant SC, Fawcett JH, et al. (2006) Biogeographic history of an Australian freshwater shrimp, Paratya australiensis(Atyidae): the role life history transition in phylogeographic diversification. Mol Ecol 15: 1083-1093.
- Santos SR (2006) Patterns of genetic connectivity among anchialine habitats: a case study of the endemic Hawaiian shrimp Halocaridina rubraon the island of Hawaii. Mol Ecol 15: 2699-2718.
- De Bruyn M, Mather PB (2007) Molecular signatures of Pleistocene sea-level changes that affected connectivity among freshwater shrimp in Indo-Australian waters. Mol Ecol 16: 4295-4307.
- Rajkumar G, Bhavan PS, Udayasuriyan R, Vadivalagan C (2015) Molecular identification of shrimp species, Penaeus semisulcatus, Metapenaeus dobsoni, Metapenaeus brevicornis, Fenneropenaeus indicus, Parapenaeopsis stylifera and Solenocera crassicornisinhabiting in the coromandel coast (Tamil Nadu, India) using MT-COI gene. Int J Fish Aqua Stud 2: 96-106.
- Umamaheswari S, Bhavan PS, Udayasuriyan R, Vadivalagan C, Kalpana R (2016) Discrimination of four marine crabs and one freshwater crab through mt-COI gene. J Entomol Zool Stud 4: 766-782.
- Bhavan PS, Umamaheswari S, Udayasuriyan R, Rajkumar G, Amritha H, et al. (2015) Discrimination of two freshwater crabs Spiralothelphusa hydrodroma and Barytelphusa jacquemontii and one mangrove crab Neosarmatium asiaticumby DNA barcoding of MT-COI gene. J Chem Biol Phy Sci 5: 1426-1440.
- Bhavan PS, Udayasuriyan R, Vadivalagan C, Kalpana R, Umamaheswari S (2016) Diversity of zooplankton in four perennial lakes of Coimbatore (India) and molecular characterization of Asplanchna intermedia, Moina micrura, Mesocyclops edax and Cypris protuberathrough mt-COI gene. J Entomol Zool Stud 4: 183-197.
- Bhavan PS, Udayasuriyan R, Kalpana R, Gayathri M (2017) Molecular identification and characterization of few crustacean zooplankton species by mt-COI gene. J Biol Nat 7: 1-23.
- Beckstead J, Augspurger CK (2004) An experimental test of resistance to cheatgrass invasion: limiting resources at different life stages. Biol Inv 6: 417-432.
- Sinclair ARE (1989) Population regulation in animals. Oxford, USA.
- Harper EB, Semlitsch RD (2007) Density dependence in the terrestrial life history stage of two anurans. Oecologia 153: 879-889.
- Kennedy BP, Nislow KH, Folt CL (2008) Habitat-mediated foraging limitations drive survival bottlenecks for juvenile salmon. Ecol 89: 2529-2541.
- Forrester GE, Steele MA (2000) Variation in the presence and cause of density-dependent mortality in three species of reef fishes. Ecol 81: 2416-2427.
- Bonenfant C, Gaillard JM, Coulson T, Festa-Bianchet M, Loison A, et al. (2009) Empirical evidence of density dependence in populations of large herbivores. Adv Ecol Res 41: 313-357.
- Loman J, Lardner B (2009) Density dependent growth in adult brown frogs Rana arvalis and Rana temporaria - A field experiment. Acta Oecol 35: 824-830.
- McMahon CR, Bester MN, Hindell MA, Brook BW, Bradshaw CJA (2009) Shifting trends: detecting environmentally mediated regulation in long-lived marine vertebrates using time-series data. Oecologia 159: 69-82.
- Elliott JM (1989) The critical period concept for juvenile survival and its relevance for population regulation in young sea trout, Salmo trutta. J Fish Biol 35: 91-98.
- Elliott JM, Hurley MA (1998) Population regulation in adult, but not juvenile, resident trout (Salmo trutta) in a lake district stream. J Anim Ecol 67: 280-286.
- Kimura M (1980) A simple method for estimating evolutionary rate of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16: 111-120.
- Xia X (2000) Data analysis in molecular biology and evolution (DAMBE).Boston, USA.
- Muse SV, Gaut BS (1994) A likelihood approach for comparing synonymous and non-synonymous nucleotide substitution rates, with application to the chloroplast genome. Mol Biol Evol 11: 715-724.
- Felsenstein J (1981) Evolutionary trees from DNA sequences –a maximum-likelihood approach. J Mol Evol 17: 368-376.
- Xia X, Xie Z, Salemi M, Chen L, Wang Y (2003) An index of substitution saturation and its application. Mol Phylogenet Evol26: 1-7.
- Xia X, Lemey P (2009) Assessing substitution saturation with DAMBE. Philippe Lemey, Marco Salemi and Anne-Mieke Vandamme. Cambridge University, USA.
- Tamura K (1992) Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C content biases. Mol Biol Evol 9: 678-687.
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870-1874.
- Vergamini FG, Pileggi LG, Mantelatto FL (2011) Phylogenetic analysis and genetic variability of the Amazon river prawn Macrobrachium amazonicum (Decapoda, Caridea, Palaemonidae) from inland and coastal waters. Contribut Zool 80: 67-83.
- Villalobos FA (1969) Problemas de especiación en América de un grupo de Palaemonidae del genero Macrobrachium. Proceedings of the World Scientific Conference on the Biology and Culture of Shrimps and Prawns. FAO Fisheries Reports 57: 1055-1066.
- Ammar D, Müller YMR, Nazari EM (2001) Biologia reprodutiva de Macrobrachium olfersii (Wiegmann) (Crustacea: Decapoda: Palaemonidae) coletados na ilha de Santa Catarina, Brasil. Revista Brasileira de Biologia 18: 529-537.
- Schwander T, Leimar O (2011) Genes as leaders and followers in evolution. Trend Ecol Evol 26: 143-151.
- Yang Z, Ranala B (2010) Bayesian species delimitation using multilocus sequence data. Proc Nat Acad Sci USA 107: 9264-9269.
- De Man JG (1879) On some species of the genus Palaemon Fabr. with descriptions of two new forms. Notes from the Leyden Museum 41: 165-184.
- Heller C (1862) Neue Crustaceen, gesammelt während der Weltumseglung der k.k. Fregatte Novara. Zweiter vorläufiger Bericht. Verhandlungen der kaiserlich-königlichen zoologisch-botanischen Gesellschaft in Wien 12: 519-528.
- Kuris AM, Ra’Anan Z, Sagi A, Cohen D (1987) Morphotypic differentiation of male Malaysian giant prawns, Macrobrachium rosenbergii. J Crust Biol 7: 219-237.
- Moraes-Riodades PMC, Valenti WC (2004) Morphotypes in male Amazon river prawns, Macrobrachium amazonicum. Aquacult 236: 297-307.
- Wortham JL, Maurik LN (2012) Morphology and morphotypes of the hawaiian river shrimp, Macrobrachium grandimanus. J Crust Biol 32: 545-556.
- Barros MP (1995) Dados biológicos sobre Macrobrachium olfersii (Wiegmann, 1836) (Decapoda: Palaemonidae) da praia da Vigia, Garopava, Santa Catarina, Brasil. Biociências 3: 239-252.
- Mossolin EM, Bueno SLS (2002) Reproductive biology of Macrobrachium olfersi(Decapoda, Palaemonidae) in São Sebastião, Brazil. J Crust Biol 22: 367-376.
- Mossolin EM, Bueno SLS (2003) Relative growth of the second pereiopod in Macrobrachium olfersi (Wiegmann, 1836) (Decapoda, Palaemonidae). Crustaceana 76: 363-376.
- Muller YMR, Nazari EM, Simões-Costa MS (2003) Embryonic stages of the freshwater prawn Macrobrachium olfersii (Decapoda, Palaemonidae). J Crust Biol 23: 869-875.
- Bate C (1868a) On a new genus, with four new species, of freshwater prawns. Proc Zool Soc UK1868: 363-368.
- Tiwari KK. 1955. Distribution of Indo-Burmese freshwater prawns of the genus Palaemon Fabr., and its bearing on the Satpura hypothesis. Bull Natl Inst Sci India 7: 230-239.
- Shokita S (1979) The distribution and speciation of the inland water shrimps and prawns from the Ryukyu Islands-II. Bull. Tokai Reg Fish Res Lab 28: 193-278.
- Jalihal DR, KN Sankolli, S Shenoy (1993) Evolution of larval developmental patterns and the process of freshwaterization in the prawn genus Macrobrachium Bate, 1868 (Decapoda, Palaemonidae). Crustaceana 65: 365-376.
- Murphy NP, Austin CM (2005) Phylogenetic relationships of the globally distributed freshwater prawn genus Macrobrachium (Crustacea: Decapoda: Palaemonidae): biogeography, taxonomy and the convergent evolution of abbreviated larval development. Zool Scr 34: 187-197.
- Beurlen K (1950) Alguns restos de crustaceous decapods dsgua doce fosseis no Brazil. Anas da Acadamia brasiliera de ciencias 22: 453-459.
- Fryer G (1977) Studies on the functional morphology and ecology of the atyid prawns of Dominica. Philos Trans Roy Soc B Biol Sci 277: 57-129.