Research Article, J Appl Bioinforma Comput Biol Vol: 12 Issue: 1
In silico Binding Affinity Studies of Antipsychotic Drugs with Protein Phosphatases by Molecular Docking
Devaraju KS*, Sarojini R Bulbule and P Aravind
Department of Biochemistry, Karnataka University, Pavate Nagar, Dharwad, Karnataka, India
*Corresponding Author: Devaraju KS
Department of Biochemistry, Karnataka University, Pavate Nagar, Dharwad, Karnataka, India
Tel: +0836-2215240;
E-mail: devarajuks@kud.ac.in
Received date: 10 January, 2023, Manuscript No. JABCB-23-67731;
Editor assigned date: 12 January, 2023, PreQC No. JABCB-23-67731 (PQ);
Reviewed date: 27 January, 2023, QC No. JABCB-23-67731;
Revised date: 03 February, 2023, Manuscript No. JABCB-23-67731 (R);
Published date: 10 February, 2023, DOI: 10.4172/2329-9533.1000249.
Citation: Devaraju KS, Bulbule SR and Aravind P (2023) In Silico Binding Affinity Studies of Antipsychotic Drugs with Protein Phosphatases by Molecular Docking. J Appl Bioinforma Comput Biol 12:1.
Abstract
Background: In eukaryotic cells, protein phosphatases have grown to be the biggest family of signaling proteins and are essential enzymes that are involved in many facets of cellular regulation. With the exception of PP1 and PP2B, PPs have not received as much research attention as kinases have. Different protein phosphatases have been targeted in clinical studies using recent meta-analyses of antipsychotic medication efficacy and tolerability. The negative effects of current standard and atypical antipsychotic medications outweigh their therapeutic benefits. Therefore, it is necessary to review their binding activity with various protein phosphatases. As a result, the current study illustrates how various phosphatases respond to the antipsychotic medications such as Clozapine, Olanzapine, and Trifluoperazine.
Method: Utilizing Maestro 9.3, three commonly used antipsychotic medications are chosen to assess the likelihood of interactions with various PPs (schrodinger). The three antipsychotic medications were filtered using Lipinski's rule (ro5) prior to docking, and these compounds were also submitted to ADME-T characteristics to look at binding interactions. The antipsychotic medications with phenothiazine groups were used in the current study due to their potential interactions with various protein phosphatases. As a result, molecular docking was used to examine the binding potential of three commonly prescribed antipsychotic medications with various protein phosphatases.
Results: The results of docking score of antipsychotic drugs with six different PP’s ranges from -7.43 kJ mol-1 to -2.91 kJ mol-1. Among all the compounds, second generation drug Olanzapine showed the highest binding affinity of -7.43 kJ mol−1 with PP5 when compared to other drugs.
Conclusion: Therefore, this study suggests that the Olanzapine has a greater affinity and exhibit non-specific modulatory effect on PP’s that need to be explored at biochemical and cellular level.
Keywords: Maestro, Protein phosphatases, Antipsychotic drugs, Clozapine, Olanzapine, Trifluoperazine, Lipinski rule, ADME-T
Introduction
There is various Protein Phosphatases (PP’s) present in the nervous system are involved in neuronal signalling [1]. In contrast to Protein Kinases (PK), PP’s dephospholyrate phospho substrate and bring about the signalling. The activity of PP’s can be either inhibitory or excitatory in nature, thereby playing an important role in Long Term Potentiation (LTP) or Long Term Depression (LTD), which is the key component of learning and memory [2]. The activity of these PP’s is to dephosphorylate any phosphosubstrate by hydrolysing phosphate group [3].
The major PP’s are classified by Ingebritsen and Cohen has been identified in the nervous system. These have been termed PP-1 PP-2A, PP-2B and PP-2C, PP-5 and PP-C based on their substrate specificity, regulation by inhibitors, and action requirements [4]. Recently, several novel membrane permeable agents have been identified which inhibit PP-1 and PP-2A. These inhibitors, which include okadaic acid, calyculin A, and tautomycin, have proven particularly valuable in studying the role of PP-1 and PP-2A in neurons [5].
PP-1 is the only enzyme in skeletal muscle with significant activity towards glycogen phosphorylase and glycogen synthase [6]. PP-1 is a major eukaryotic protein serine/threonine phosphatase that regulates an enormous variety of cellular functions through the interaction of its catalytic subunit (PP1c) with over fifty different established or putative regulatory subunits [7]. Reversible protein serine/threonine phosphorylation is a significant component of the intracellular signaling machinery, directing such diverse functions as neurotransmission, muscle contraction, glycogen synthesis, T-cell activation, neuronal plasticity and cell proliferation [8].
PP-2A comprises a family of serine/threonine phosphatases, minimally containing a well conserved catalytic subunit, the activity of which is highly regulated. Regulation is accomplished mainly by members of a family of regulatory subunits, which determine the substrate specificity, sub cellular localization and catalytic activity of the PP-2A holoenzymes [9]. It involves in glycogen metabolism, Hepatic Metabolism, glycolysis/gluconeogenesis, fatty acid synthesis, and amino acid breakdown.
PP-2C was originally identified as an Mg2+ dependent protein phosphatase (half maximal activity at 1 mM Mg2+). The enzyme is monomeric with a molecular mass of 43-48 kDa two major isoforms (A, B) of PP-2C have been cloned [10]. Further analysis revealed the existence of subtypes for both isoforms. The latter observation might be important, since PP-2C was suggested to play a role in Ca2+ dependent signal transduction in brain. More specifically, PP-2C might catalyze the dephosphorylation of autophosphorylated Ca2+/ calmodulin protein kinases II the granule cells of cerebellum [10]. It accounts a very small portion in hepatic glycogen synthase. PP-2C is vitally involved in Abscisic Acid (ABA) signaling [11].
PP-5 enzyme is predominantly nuclear, but is apparently excluded from the nucleolus. PP-5 is related to PP-l, PP-2A and PP-2B, but has an N terminal extension of 200 amino acids. This extension contains Tetratricopeptide Repeat (TPR) motifs, also found in several other nuclear proteins.
PP-7 it regulates dephosphorylation of Mitogen Activated Protein Kinases (MAPKs) plays a key role in determining the magnitude and duration of kinase activation and hence the physiological outcome of signaling [12].
A recent meta analysis of antipsychotic drug efficacy and tolerability has been used to design and target different PP’s in clinical trials. Existing typical and atypical antipsychotic drugs have paved more of side effects than their treatment. Therefore, the current study, the antipsychotic drugs containing phenothiazine groups was used for their plausible interaction with different PP’s by in silico approach. Therefore, three majorly prescribed antipsychotic drugs i.e. Clozapine (CLZ), Olanzapine (OLZ) and Trifluoperazine (TFP) were selected to analyze their binding potential with different PP’s using molecular docking i.e. Maestro 9.3 (Schrodinger, LLC, New York).
In recent years CLZ is an atypical antipsychotic drug which has extraordinary pharmacological profile with potent interactions at a large number of, mainly post synaptically localized, biogenic amine receptors, including several serotonin, adrenergic, and muscarinic G Protein Coupled Receptor (GPCRs) [13-16]. CLZ also demonstrates high affinity for the 5-HT2C, 5-HT6 and 5-HT7 receptor [17]. CLZ binds to D4 receptor with a higher affinity (10 times higher than to D2 and D3 receptor) [18].
OLZ is an atypical antipsychotic drug and is approved by the US food and drug administration for the treatment of schizophrenia and bipolar disorder. It has stronger affinity for the 5-HT2 muscarinic and histamine receptor than for dopamine D2 receptor, is known to cross the membrane and concentrates intracellularly [19,20]. OLZ binds to the 5-HT2 serotonin receptor and D2 dopamine receptor which are key for maintaining chemical balance within the brain. When a patient has schizophrenia, these receptor start malfunctioning and thus creating the chemical imbalance in the brain. OLZ function is to prevent these receptors from further binding them in such a way that they stop working. It’s anticholinergic, antihistaminic and alpha adrenergenic blocking activity contributes to its therapeutic and adverse action that the drug performs, thus producing fewer negative effects than most antipsychotics on part of brain designed for motor skills and coordination of movement.
On the other hand TFZ is a typical antipsychotic drug, is an antagonist of Calmodulin (CaM), an essential regulator of calcium dependent signal transduction. It blocks dopamine D1 and D2 receptor in the mesocortical and mesolimbic pathway. TFZ has central antiadrenergic, anti-dopaminergic effect. TFZ causes extrapyramidal side effects akathisia, dystonia and Parkinsonism and also causes anticholinergic side effects such as red eye xerostomia (dry mouth).
Indeed different PP’s play a very important role in various molecular events as described above and finally reaching out on a biological/physiological response in the nervous system. CaN is inhibited by the TFZ and also with Auto Inhibitory Domain (AID). However a series of peptides generated from AID of CaN, are used for binding affinity studies by molecular modeling. In our earlier investigation through molecular docking we were able to demonstrate CaN an exhibits binding affinity with various molecules that are associated with the normal molecular mechanisms. CaN an exhibits binding affinity with FK506, Cyclosporin A, TFZ, FKBP12, CyPA and so on. In addition, our group also designed phosphopeptide derived from PKA subunit type II for assay of CaN and also developed assay method for CaN. Hence in the current study, is to investigate the plausibility of binding affinity of the few antipsychotic drugs on different PP’s by molecular docking approach using Maestro 9.3 (Schrodinger, LLC, New York). This study emphasis comparison of second generation antipsychotic drugs i.e. CLZ and OLZ with firstgeneration drug TFP for their binding affinity.
Materials and Methods
Tools and servers used
3D structures of antipsychotic drugs molecules were extracted from Pubchem. The protein structural files were fetched out from PDB as shown in (Table 1). Protein-ligand interactive visualization and analysis was carried out Maestro. ADME-T properties of molecules were identified using QikProp in schrodinger. Molecular docking was performed using Maestro 9.3.
Table 1: 3D structures model of different PP’s retrieved from Protein data bank with their PDB Id.
Prediction of drug likeness properties
Prior to optimization of compounds, CLZ, OLZ and TFZ compounds were filtered based on drug likeness properties. The evaluation of drug likeness was performed on the basis of Lipinski’s rule of five (ro5). The parameters were calculated using QikProp 4.4 (Schrödinger, LLC, and New York). Criteria for ro5 include H-Bond Donors (HBD ≤ 5), H-Bond Acceptors (HBA ≤ 10), molecular weight (MW<500), and the octanol/water partition coefficient (log P<5).
Prediction of ADME-T properties of the selected ligands
ADME-T properties were predicted for 11 molecular descriptors available with QikProp. The selected molecules were assessed for pharmacokinetic properties like solubility, confirmation independent predicted aqueous solubility, blood brain barrier permeability, metabolic reactions, Jorgensen’s rule, central nervous system activity, brain/blood partition coefficient, skin permeability, transdermal transport rate, human serum albumin and Herg K+. Based on the results, each ligand was assigned with respective drug likeness and drug score.
Preparation of macromolecule
The structures were prepared and refined using the Protein preparation wizard of Schrödinger Maestro. Charges and bond orders were assigned, hydrogen’s were added to the heavy atoms and all waters molecules were deleted. Using force field OPLS-5, minimization was carried out by setting the maximum heavy atom root mean square division (RMSD) to 0.30 A??. Later it was added with Cterminal oxygen, polar hydrogen and gasteiger charges.
Preparation of inhibitors
The antipsychotics drugs CLZ, OLZ and TFP were retrieved from PubChem (Figure 1). These ligands were prepared using the Lig prep module of Schrödinger Maestro. Torsions of the ligands were modified and assigned with appropriate protonation states. In glide (Schrödinger), 32 stereo chemical structures were generated per ligand with possible states at pH 7.0 ± 2.0. It was optimized by ionizer, tautomerized, desalted and optimized by producing low energy 3D structure for the ligand under the OPLS-5 force field while retaining the specified chiralities of the input Maestro file (Table 2).
Table 2: Antipsychotic drugs retrieved from PubChem.
Figure 1: Structure of Clozapine. (A) (Pubchem Id-135398737), Olanzapine; (B) (Pubchem Id-135398745) and Trifuloperazine; (C) (Pubchem Id-5566).
Molecular docking
It helps in predicting the intermolecular framework formed between a protein and a small molecule or a protein and protein and suggest the binding modes responsible for inhibition of the protein. To precisely carry out docking studies one requires the high resolution X-ray, NMR or homology-modeled structure with known/predicted binding site in the biomolecule.
Receptor grid generations
Receptor grids were calculated for prepared proteins such that various ligand poses bind within the predicted active site for docking. In Glide, grids were generated keeping the default parameters of Vander Waals scaling factor 1.00 and charge cut off 0.30 subjected to OPLS 2001 force field. A cubic box of specific dimensions centered on the centroid of the active site molecular residues was generated for each receptor. The grid box was set to 14 A?? x 14 A?? x 14 A?? for all docking experiments.
Extra precision
Extra precision was performed using the induced module of Schrödinger-Maestro. The entire receptor molecule was minimized with an RMSD cutoff of 0.18 A??. It was selected for generation of the centroid of the residues and the box size was generated automatically. The initial Glide docking for each ligand was carried out, with a receptor and Vander Waals scaling of 0.70 A?? and 0.50 A?? respectively. The numbers of poses generated were set to be 20. Prime side chain prediction and minimization were carried out in which residues were refined within 5.0 A?? of ligand poses and side chains were optimized to a ligand structure and conformation that is induced fit to each pose of the receptor structure.
Results
Lipinski rule or rule of five (ro5) was applied to evaluate drug like properties of the designed inhibitors. A value of log P not greater than 5, hydrogen bond donors not more than 5, hydrogen bond acceptors not greater than 10 and molecular weight not exceeding 500 Da are generally considered for an orally bioavailable and drug like compound. Interestingly, none of the inhibitors disobeyed Lipinski’s rule of five (Table 3). This is a good proof that the designed inhibitors would be orally bioavailable and pharmacologically active as potential antipsychotic drugs.
Table 3: Drug like properties of three antipsychotic drugs CLZ, OLZ and TFZ as inhibitors.
The three compounds were analyzed for their ADME-T properties using QikProp 4.4 tool of maestro software to determine their safety profile. The compounds show significant values for ADME-T properties that one analyzed and exhibited drug-likeness properties (Table 4). The compounds have QikProp parameters within the acceptable range, except for QPPCaco and QPlogHERG parameters. QPPCaco parameter is used to assess the permeability of compounds across the gut-blood barrier. CLZ and OLZ showed excellent permeability and TFZ have intermediate permeability across the gut blood barrier. Human Ether à go go Related Gene (hERG) parameter is used to determine the potential cardiac toxicity of the compounds. The hERG encodes a potassium ion (K+) channel which plays a role during systolic and diastolic activities of the heart. The blockage of hERG K+ channels can lead to cardiac arrythmia. CLZ and TFZ compounds have logIC50 (hERG) values more than the acceptable range for the blockage of hERG K+ channels (logIC50 (hERG)<−5); however, the values are near the borderline. Thus, these compounds were proceeded for further analysis.
Where, QPlogS: denotes aqueous solubility; CIQPlogS: Conformation Independent predicted aqueous solubility; QPPCaco: Predicted apparent gut-blood barrier permeability; #metab: Number of likely metabolic reaction; ro3: The number of violations of Jorgensen’s rule of three; CNS: Central Nervous System activity; QPlogBB: Brain/blood partition coefficient; QPlogKp: Predicted skin permeability; Jm: Predicted maximum transdermal transport rate; QPlogKhsa: Prediction of binding human serum albumin; QpHERG: Predicted IC50 value for blockage of HERG K+ channels.
Table 4: Predicted ADME-T values of CLZ, OLZ and TFZ using a QikProp module of Schrodinger.
Furthermore, in order to understand the possible interaction of the antipsychotic drugs for different PP’s, molecular docking studies were performed on different PP’s using Glide software of Schrodinger molecular docking tool kit to evaluate the binding affinity and the interaction between the antipsychotic drugs and different PP’s. The way of combined ligand and structure based approach was used to analyze the binding affinity of enzyme and its interaction with their inhibitors. The docking score of the antipsychotic drug varies from -7.43 kJ mol-1 to -2.91 kJ mol-1 (Table 5).
Table 5: Binding free energy (ΔG) of different inhibitors with PP’s in –kJ mol-1.
Amongst the three drugs, OLZ showed the highest binding affinity with PP-5 (-7.43 kJ mol-1) followed by PP-1 (-6.74 kJ mol-1) and PP-2B (-6.61 kJ mol-1). The evdv and ecoul are the predicted Vander Waal’s interaction and electrostatic interaction energies between the ligand and the protein docking results clearly indicate that the water molecule present in the active site of an enzyme PP-5, PP-1 and PP-2B has a less important role in binding with OLZ (Table 6).
Table 6: Molecular docking results of OLZ with PP-5, PP-1, and PP-2B.
All the target compounds were docked at the binding site of PP’s. The docking pictures (Figure 2). The binding site comprises of polar and hydrophobic residues which involve in the binding with the target. The substituent ring with the two nitrogen’s in a 1,4 arrangements is a piperidine ring, which is surrounded by a hydrophobic pocket of the PP-5.
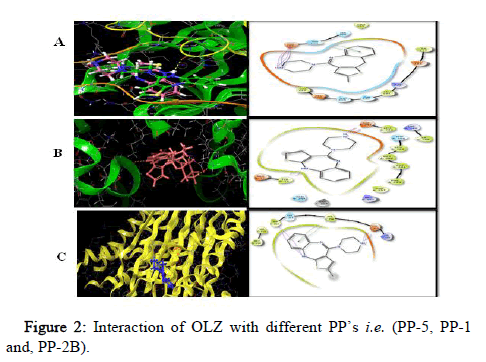
Figure 2: Interaction of OLZ with different PP’s i.e. (PP-5, PP-1 and, PP-2B).
PP’s (Figure 2) show as a green ribbon and OLZ in five different colors depicting the atoms present in it; white=hydrogen, blue=nitrogen, red=oxygen, grey=carbon and yellow=sulfur bound to the enzyme active site. It indicates the 3D structures of three different PP’s i.e. PP-5, PP-1 and PP-2B docked with the inhibitor OLZ.
PP5 (Figure 2) forms one hydrogen bond i.e. hydrogen of OLZ to oxygen of Asp 295 at 1.65 A°, one salt bridge from nitrogen of OLZ to oxygen of Asp 295 at 4.49 A° and π-π interaction from benzene ring of OLZ to imidazole ring of His 296 in chain A. PP-1 is depicted in green ribbon and OLZ in olive red, with one hydrogen bonding from hydrogen of OLZ to oxygen of met 329 in chain A at 2.33 A° whereas PP-2B is depicted in yellow complexed with the inhibitor OLZ in blue which forms two hydrogen bonds from hydrogen of OLZ to oxygen of Glu 359 at 1.96 A° and Trp 352 at 1.96 A°, and π-π interaction was from benzene ring of OLZ to phenyl ring of Phe 356 in Chain C, salt bridge is formed from nitrogen of OLZ to oxygen of Glu 359 at 3.18 A °, aromatic bond was formed from hydrogen of OLZ to oxygen of Met 118 in chain D at 2.46 A°.
The docking calculations were performed using IFD of Schrödinger-Maestro. The left panel 2D interactions of the ligand OLZ with three different PP’s. Interactions between the best pose found of the OLZ with PP-5, PP1, and PP2B. Docking of OLZ into the active site of PP’s with key amino acid residues (colored by green, blue red and light blue) in represented ligand binding poses. The green ribbon represents the hydrophobic, whereas the blue ribbon represents positively charged, red ribbon represents negatively charged and light blue represents the polar nature of the molecule respectively (Figure 3).
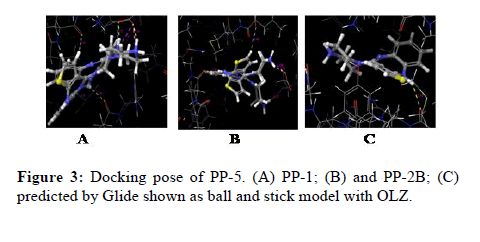
Figure 3: Docking pose of PP-5. (A) PP-1; (B) and PP-2B; (C) predicted by Glide shown as ball and stick model with OLZ.
The binding energy was observed in the range of -7.43 kJ mol-1 to -2.91 kJ mol-1 (Figure 4).
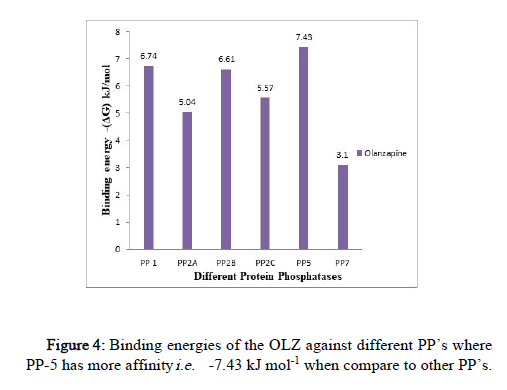
Figure 4: Binding energies of the OLZ against different PP’s where PP-5 has more affinity i.e. -7.43 kJ mol-1 when compare to other PP’s.
The highest binding affinity of OLZ is exhibited towards PP-5 of (-7.43 kJ mol-1) ≥ PP-1 (-6.74 kJ mol-1) ≥ PP-2B (-6.61 kJ mol-1) when compared to other PP’s; the effect was exhibited by PP-2C (-5.57 kJ mol-1), PP-2B (-5.04 kJ mol-1) and PP-7 (-3.1 kJ mol-1). (The docking results of other PP’s against different antipsychotic drugs are reported in a histogram as supplementary data).
Discussion
Ro5 was used to evaluate the oral availability of the CLZ, OLZ and TFZ. Development of orally active drug give several advantages in delivering drugs to the patients such as safe, good patient compliance, ease of ingestion, pain avoidance and versatility to accommodate various types of drugs 40. In addition, ro5 is widely used in drug development process to narrow the number of compounds from large library before virtual screening campaign. According to Lipinski and his colleagues, the rule was formulated based on analysis of four consistent physicochemical properties of 2,245 oral based drugs in the World Drug Index (WDI) database. These properties are molecular weight (MW) is ≤ 500 Dalton (Da), the octanol/water partition coefficient (logP) is <5, the number of H-bond (HBD) is ≤ 5, and the number of H Bond Acceptors (HBA) is ≤ 10. Later, the criteria were improved by adding Number of Rotatable Bonds (NRB) because it plays a role in drug metabolism and pharmacokinetics profiles.
Virtual screening, in silico ADME and molecular docking studies were carried out to identify potential inhibitors for PP’s. The Pharmacokinetic parameters such as aqueous solubility (QPlogS), CIQPlogS: Conformation independent predicted aqueous solubility, QPPCaco: Predicted apparent gut-blood barrier permeability, #metab: Number of likely metabolic reaction, ro3: The number of violations of Jorgensen’s rule of three, CNS: Central nervous system activity, QPlogBB-Brain/blood partition coefficient, QPlogKp: Predicted skin permeability, Jm: Predicted maximum transdermal transport rate, QPlogKhsa: Prediction of binding human serum albumin and QpHERG-predicted IC50 value for blockage of HERG K+ channels were calculated. The corresponding values are noted in Table 4 for all the three antipsychotic drugs. The pharmacokinetic properties of CLZ, OLZ and TFZ exhibited that the values were within acceptable range except for QPPCaco and QPlogHERG parameters; QPPCaco parameter for CLZ and OLZ showed excellent permeability and TFZ have intermediate permeability across the gut-blood barrier. Human Ether à go go Related Gene (hERG) parameter for CLZ and TFZ compounds have logIC50 (hERG) values more than the acceptable range for the blockage of hERG K+ channels. Hence OLZ would be potential antipsychotic drug for PP’s.
Molecular docking is a widely used, relatively fast, and economical computational tool for predicting in silico the binding modes and affinities of molecular recognition events and the concept of free energy (ΔG) is used to determine the binding affinity of protein-ligand complex in docking studies. The negative or low value of ΔG indicates the strong binding affinity between protein-ligand complexes. Hence, in the current study, the binding affinity was carried out and the determined binding free energy as shown in Table 3 which reflects the binding affinity of the different ligands to a PP’s by calculating intermolecular and torsional free energies using Maestro.
More recently the antipsychotic drug TFZ has been used extensively to probe for CaM dependent reactions. It is a phenothaizine derivative antipsychotic drug being observed as a PP-2B inhibitor. TFZ has shown the highest binding affinity towards PP-1 (-6.38 kJ/mol) when compared to PP-2B (-6.14 kJ/mol) and PP-2A (-6.07 kJ/mol) followed by PP-2C (-6.01 kJ/mol) and PP-7 (- 2.91 kJ/mol). Free energies of TFZ binding to Chain A of PP-2B ranged from -7.2 to -6.9 kcal/mol, while -7.8 to -7.2 kJ/mol were observed in TFZ binding to Chain A of PP-2B using Maestro (Schrodinger, New York).
However there is less/no study being carried out investigate the other popular antipsychotic drugs such as CLZ and OLZ with PP’s and also TFZ with other PP’s. In the present study, phenothiazine derivatives such as CLZ, OLZ, and TFZ were docked against different PP’s. Results indicate the relative binding affinity of phenothiazine derivatives i.e. OLZ with different PP’s and mainly concentrating on PP-5, PP-1, and PP-2B.
CLZ, a second generation antipsychotic drug strongly inhibits PP-2B activity, but had minimal effect on PP-1 and PP-2A activities at the concentrations studies. Therefore, docking studies of CLZ with different PP’s show that CLZ has higher binding affinity with PP- 2C (-5.91 kJ/mol) when compared to PP -1 (-5.63 kJ/mol) and PP-5 (-5.05 kJ/mol) followed by PP-2B (-4.52 kJ/mol) and (PP-2A - 4.05 kJ/mol) and has least binding affinity with PP-7 (-3.44 kJ/mol).
Results obtained indicate that the presence of piperidine ring in OLZ makes stronger activator in comparison with diazepine rings. In addition to diazepine rings, the piperidine rings have shown significant binding affinity when compared to other moieties. On this basis, some of the pharmacophoric features were generated which included hydrogen and halogen bonding, salt bridges, aromatic bonds, π-cation and π-π interactions.
OLZ has the highest binding affinity towards PP-5 (-7.43 kJ/mol), when compared to PP-1 (- 6.74 kJ/mol) and PP- 2B (-6.61 kJ/mol), followed by PP-2C (-5.51 kJ/mol) and PP-2A (-5.04 kJ/mol) and least affinity towards PP-7 (-3.15 kJ/mol). As shown in Figure 2A the active site residues of PP-5 protein include Glu, Asp, Glu, Thr and His. In Figure 2B, the active site residues of PP-1 include Asp, Tyr, and Thr. In Figure 2C Glu, Gln, Met, Tyr and His were observed interacting with the drug molecule illustrating the hydrophobic nature of PP’s at their active sites. The docking score for different PP’s nearly range from -7.43 kJ mol-1 to -3.1 kJ mol-1 which clearly indicates that the second generation drug OLZ has more binding affinity compared to other drugs with PP’s.
The present study is done to predict the binding affinity of antipsychotic drugs with different PP’s in a view to discover potential therapeutic strategy in silico approach which can provide significant insights into other major characteristics of antipsychotic drugs and their binding with the PP’s. Henceforth, the results obtained would lead us to know the different roles of antipsychotic drugs and their interactions with different PP’s. This can be further used as potential targets in the field of drug designing.
Conclusion
In this work, drug likeness properties, in silico ADME-T and molecular docking studies were carried out to identify potential inhibitors of PP’s. Three compounds satisfied drug likeness properties based on ro5. OLZ showed the highest potential in this work. With respect to the reference drug, TFZ (FDA approved drug), OLZ showed a lower XP GScore of −7.43 kcal/mol (TFZ: −3.41 kcal/mol). QPlogHERG parameter for OLZ was −5.815, which is compounds have logIC50 (hERG) values more than the acceptable range for the blockage of hERG K+ channels. In conclusion, this study predicts that the binding affinity of three antipsychotic drugs CLZ, OLZ and TFZ found to be important, whereas OLZ was found to be a promising antipsychotic drug that has higher binding affinity with inhibits PP-5 activity strongly. It showed a higher binding energy of -7.43 kJ mol-1 towards PP-5 compared to the other two antipsychotic drugs. However, further in vitro and in vivo studies would help us in recognizing the potentiality of the antipsychotic drug in inhibition of PP’s.
References
- Tweedie-Cullen, R.Y., Park, C.S., Mansuy, I.M. (2011). Protein Phosphatases in the Brain: Regulation, Function and Disease. In: Vidal, C. (eds) Post-Translational Modifications in Health and Disease. Protein Reviews, Springer, New York, PP.233-257.
- Winder DG, Sweatt JD (2001) Roles of serine/threonine phosphatases in hippocampel synaptic plasticity. Nat Rev Neurosci 2:461-474.
- Szöör B (2010) Trypanosomatid Protein Phosphatases. Mol Biochem Parasitol 173:53–63.
[Crossref] [Googlescholar] [Indexed]
- Ingebritsen TS, Cohen P. The protein phosphatases involved in cellular regulation: 1. Classification and substrate specificities. Eur J Biochem 132:255-261.
[Crossref] [Googlescholar] [Indexed]
- Swingle M, Ni L, Honkanen RE (2009) Small Molecule Inhibitors of Ser/Thr Protein Phosphatases: Specificity, Use and Common Forms of Abuse. Methods Mol Biol 15.
- Cohen P (1989) The structure and regulation of protein phosphatases. Ann Rev Biochem 58:453-508.
- Cohen P (2002) Protein kinases--the major drug targets of the twenty-first century?. Nat Rev Drug Discov 20 1:309-315.
[Crossref] [Googlescholar][Indexed]
- Cohen PT (2002) Protein phosphatase 1–targeted in many directions. J cell Sci 115:241-256.
[Crossref] [Googlescholar][Indexed]
- Janssens V, Goris J (2001) Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem J 353:417-439.
[Crossref] [Googlescholar] [Indexed]
- Wera S, Hemmings BA (1995) Serine/threonine protein phosphatases. Biochem J 311:17-29.
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324:1068-1071.
[Crossref] [Googlescholar] [Indexed]
- Owens DM, Keyse SM (2007) Differential regulation of MAP kinase signalling by dual-specificity protein phosphatases. Oncogene 26:3203.
- Meltzer HY, Bastani B, Ramirez L, Matsubara S (1989) Clozapine: new research on efficacy and mechanism of action. Eur Arch Psychiatry Neurol Sci 238:332-339.
- Enna SJ, Bennett Jr JP, Bylund DB, Creese I, Burt DR, et al. (1997) Neurotransmitter receptor binding: regional distribution in human brain. J Neurochem 28:233-236.
- Bolden C, Cusack BE, Richelson E (1992) Antagonism by antimuscarinic and neuroleptic compounds at the five cloned human muscarinic cholinergic receptors expressed in Chinese hamster ovary cells. J Pharmacol Exp Ther 260:576-580. [Crossref]
- Roth BL, Craigo SC, Choudhary MS, Uluer A, Monsma FJ, et al. (1994) Binding of typical and atypical antipsychotic agents to 5-hydroxytryptamine-6 and 5-hydroxytryptamine-7 receptors. J Pharmacol Exp Ther 268:1403-1410.
- Horacek J, Bubenikova-Valesova V, Kopecek M, Palenicek T, Dockery C, et al. (2006) Mechanism of Action of Atypical Antipsychotic Drugs and the Neurobiology of Schizophrenia. CNS Drugs, 389-409.
[Crossref] [Googlescholar][Indexed]
- Van Tol HH, Bunzow JR, Guan HC, Sunahara RK, Seeman P, et al. (1991) Cloning of the gene for a human dopamine D4 receptor with high affinity for the antipsychotic clozapine. Nature 350:610.
[Crossref] [Googlescholar][Indexed]
- Shitij Kapur MP (1998) 5-HT2 and D2 Receptor Occupancy of Olanzapine in Schizophrenia: A PET Investigation. Am J Psychiatry , 921–928.
- Loryan I, Melander E, Svensson M, Payan M, König F, et al. (2016) In-depth neuropharmacokinetic analysis of antipsychotics based on a novel approach to estimate unbound target-site concentration in CNS regions: link to spatial receptor occupancy. Mol Psychiatry 21:1527-1536.
[Crossref] [Googlescholar] [Indexed]

