Research Article, J Appl Bioinforma Comput Biol Vol: 9 Issue: 5
In Silico Molecular Docking Study of Some Already Approved Antiviral Drugs, Vitamins and Phytochemical Components Against Rna Dependent Rna Polymerase of Sars-Cov-2 Virus
Md. Nabil, Rafat Alam*, Quazi Ashiqur Rahman, Md.Mahabub UL islam and Md. Ali Asif Noor
North South University, Bashundhara, Dhaka 1229, Bangladesh
*Corresponding Author: Rafat Alam
North South University, Dhaka, Bangladesh
E-mail: rafat.alam@northsouth.edu
Received: July 31, 2020 Accepted: September 30, 2020 Published: October 07, 2020
Citation: Nabil Md, Alam R, Rahman QA, Islam MMUL, Noor Md AA. (2020) In Silico Molecular Docking Study of Some Already Approved Antiviral Drugs, Vitamins and Phytochemical Components against RNA Dependent RNA Polymerase of Sars-Cov-2 Virus. J Appl Bioinforma Comput Biol 9:5.. doi: 10.37532/jabcb.2020.9(5).182
Abstract
Background: Coronavirus Disease 2019 (COVID 19) is a disease caused by Sars-Cov-2 virus which has been announced as a pandemic and a global emergency health concern by WHO. It has already affected millions of people across the world. A lot of people have succumbed to this virus as well. The virus was originated in a small town of China (Wuhan) in December, 2019 and spread throughout the world very fast. A number of research works have been conducted to find an effective treatment for this deadly disease. The invention of a medicine through the conventional way against a virus is huge time consuming and extravagantly costly. Therefore, we have conducted a molecular docking study with a number of existing antiviral drugs available in the market for the treatment of different viral diseases, two vitamins (Vitamin C and D) and phytochemical components (Thymoquinone and Carvacrol, two major constituents of Nigella sativa) in search of an effective inhibitor of RNA dependent RNA polymerase enzyme of Sars-Cov-2 virus.
Materials and Methods: Molecular docking was conducted using the software Autodock Vina which is available in the MGLtools website. We also analyzed the amino acid interactions of the ligands by using Discovery Studio.
Results & Conclusion: Out of 20 ligands we sorted out Paritaprevir, Elvitegravir, Ledipasvir, Ribavirin and Favipiravir as top five ligands based on the binding affinity, ( -9.7, -6.5, -9.4, -5.8 and -5.8 KJ/mol respectively) RMSD values (2.19, 2.05, 2.24, 1.93 and 2.14 respectively) and amino acid interaction patterns (all five ligands possessed considerable number of interactions with hydrophobic and hydrogen bonds without any unfavorable bumps) to suggest for further study in pursuit of finding an inhibitor of RNA dependent RNA polymerase enzyme of Sars-Cov-2 virus.
Keywords: COVID-19; Molecular Docking; Antivirals; RMSD Value; Docking Interactions
Introduction
Coronavirus Disease 2019 (COVID-19), causes by a strain of coronavirus is a highly transmissible disease that belongs to the family of Coronaviridae in the Nidovirales order. It has been named ‘Coronavirus’ due to the presence of crown like spikes on the outer surface of the virus. Previously, symptoms such as acute lung injury & acute respiratory distress syndrome (ARDS) used to result in due to the presence of severe acute respiratory syndrome coronavirus (SARSCoV), H5N1 influenza A, H1N1 2009 and Middle East respiratory syndrome coronavirus (MERS-CoV), leading to pulmonary failure & results in fatality. Symptoms such as respiratory and intestinal infections were not considered to be pathogenic [1].
It was thought to infect only animals before experiencing the outbreak of SARS-CoV, back in 2002, in Guangdong, China. In most of the cases, coronavirus transmitted before that time caused mild infections in immunocompetent people [1]. The virus was also detected as a beta coronavirus group as it exhibited pneumonia symptoms with diffused alveolar injury. Decade later, MERS-CoV emerged in Middle Eastern countries, becomes epidemic in the region. A couple of Saudi Arabian nationals were detected to be infected by it. WHO reported that MERS-CoV infected more than 2428 individuals and 838 deaths.The infection of this virus initiated from the mild upper respiratory injury, as the progression leads to severe respiratory disease. Similarities with the SARS-Coronavirus is that MERS infected patients suffer from pneumonia, followed by ARDS and renal failure. Both SARS-CoV & MERS-CoV have been detected as a Coronavirus. At the end of 2019, WHO got information about several cases of pneumonia having unfamiliar symptoms, which was identified as the symptoms of novel coronavirus. Also, Chinese Center for Disease Control & Prevention and Wuhan city health authorities reported outbreak of pneumonia with unknown cause back in 31st December, 2019 [2].
One of the emerging business town of China, known as Wuhan, experienced the outbreak of this virus, killed almost 3000 people in China. Chinese researchers named the virus as 2019-nCoV, which is thought to be initiated from the Hunan seafood market in the Wuhan city of China, killed more than 1800 and infected over seventy thousand individuals within the first fifty days of epidemic.This report represents the spreading ability of the virus. SARS-CoV-2 was named by The International Taxonomy of Virus (ICTV) as well named the disease as COVID-19. Mortality rate is also higher, as SARS-CoV infected more than 215 countries, whereas novel coronavirus infected more than over 16.7 Million with the death toll over 661 Thousand [3]. This statistics showed that transmission rate is higher in SARSCoV- 2 compared to SARS-CoV. Many factors can be responsible for the speedy transmission of SARS-CoV-2 including S protein having genetic recombination in the RBD region of SARS-CoV-2, enhancing the transmission ability of SARS-CoV-2 [4].
Source of Transmission
In 2001, samples were collected from some individuals in Hong Kong. According to their molecular assessment, 2.5% frequency rate of antibodies were found against SARS-Coronavirus. These suggested the existence of SARS Coronavirus in human being before finally being identified in 2003. Later on, source of replication was found as Rhinolophus bats having anti-SARS-CoV antibodies. It was then considered as the source of replication. In case of MERS-Coronavirus, camels were found as a primary host. In recent studies, this MERSCoronavirus have also been detected in two types of bats: Pipistrellus and Perimyotis, suggesting that bats are the key host and transmitting medium of the virus [4].
Clinical Features of COVID-19
Some of the transmission analysis report suggests that fever and respiratory symptoms appeared 3-7 days after exposure to the virus along with the symptoms of dry cough and fatigue. In rare cases, nasal congestion, rhinorrhea, sore throat & myalgia were noticed. Other symptoms like palpitation, diarrhea or headache were also noticed [2].
Virology of SARS-CoV-2
Coronavirus are enveloped virus with a positive single stranded RNA genome. It has a size range of 65-125 mm in diameter and contains a single-stranded RNA as a nucleic material, ranging the size from 26-32 kbs in length. Identified subgroups of Coronavirus include: α, β, γ, δ with human coronavirus being detected in the α Coronavirus (HCoV-229E and NL63) & β Coronavirus (MERS-CoV, SARS-CoV, HCoV-OC43 and HCoV-HKU1). It was found to be the member of β group of coronavirus. Further analysis of homologous recombination reveals that receptor binding spike glycoprotein of novel coronavirus has been developed from the SARS-CoV and an unknown beta-CoV. It has been found that all coronavirus contains specific genes in ORF1 downstream regions, encoding protein for viral replication. Spikes made of glycoprotein, located on the outer surface are mainly responsible for the attachment and entry of the virus to host cells. Due to the presence of receptor binding domain (RBD), which are loosely attached among virus, infects the multiple hosts. Other coronavirus mainly recognizes aminopeptidases or carbohydrates as a main receptor, act as the entrance inside the human cell, whereas SARS-CoV & MERS-CoV recognizes exopeptidases. [5]. Entrance mechanism of coronavirus mainly depends upon different cellular proteases including human airway trypsin like protease (HAT), cathepsins & transmembrane protease serine 2 (TMPRSS2), acts to split spike proteins, establish changes in penetration pattern [6,7]. For the activity, MERS-Coronavirus requires Dipeptidyl Peptidase 4 enzyme (DPP-4), as the receptor binding S1 protein of the MERS-CoV spike protein occupied with DPP4, mainly being lysated from susceptible Huh-7 cells. Also, MERS-CoV binds to the DPP4 from multiple species promoting the transmission to humans & other species. As a receptor, SARS-coronavirus requires Angiotensin-converting enzyme (ACE2). Besides having spike protein, SARS-CoV-2 also have other polyproteins, nucleoproteins & membrane proteins, including RNA polymerase, 3-chymotrypsinlike protease, papain-like-protease, helicase, glycoprotein as well as accessory proteins. Van der Waals force being maintained as the spike protein of SARS-CoV-2 contains 3D structure in the RBD region [4]. Besides this, two-thirds of the RNA of coronavirus have encoded viral polymerase, RNA synthesis materials and two large nonstructural proteins, not playing the role as a host response modulation. SARSCoV & MERS-CoV contains two polyproteins known as pp1a & pp1ab. Pp1ab produces 15 non-structural proteins (nsp1-nsp10 and nsp12-nsp16). Despite the mechanism is not being fully understood, np3 plays the role in auto proteolytic cleavage, whereas nsp12 plays its role in RNA-dependent RNA-polymerase [8,9]. The role of nonstructural proteins is to rearrange membranes, which is originated from the rough ER, gets converted into double membrane vesicles, which is the source of viral replication & transcription [4]. Adaptation to the human host occurs when genome encoding occurs inside the cell, facilitates gene expression & encodes useful accessory proteins [10].
Coronavirus Entry & Replication
Coronavirus S protein plays an important role of determining virus entry into the cell [11]. The envelope spike glycoprotein binds to ACE2 receptor for SARS-CoV & SARS-CoV-2 [12] CD209L for SARS-CoV, DPP-4 for MERS-CoV. Direct membrane fusion between the virus & plasma membrane allows the entry of SARS-CoV-2 into cells. Also, critical proteolytic cleavage event occurred at S protein of SARS-CoV-2 at S2’ position, lets to the mediation of membrane fusion & viral infectivity. MERS-CoV evolved with the membrane fusion which requires abnormal two step furin activation [13]. Viral RNA genome is released into the cytoplasm after the virus enters the cell and gets translated into two polyproteins & structural proteins, after which replication of viral genome started [14]. Also, series of process occurs as the newly formed envelope glycoproteins are inserted into the membrane of the ER/ Golgi, combination of genomic RNA & nucleocapsid protein helps to form nucleocapsid & viral particles germinated into the ER-Golgi intermediate compartment (ERGIC). Then the virus gets released as the vesicles containing the virus particles fused with the plasma membrane [15,16].
Presentation of Antigen in Coronavirus Infection
After virus enters the cell, antigen is presented to the antigen presenting cells (APC), acting as a central part of the body’s anti-viral immunity. Despite having lack of report about it, some information from previous report researches on SARS-CoV & MERS-CoV have shown that the antigen presentation of SARS-CoV mainly depends on MHC I molecules [17], MHC II also plays role in the presentation. Previous reports have also shown relation between HLA polymorphs and the susceptibility of SARS-CoV, such as HLA-B*4601, HLA-B*0703, HLA-DR B1*1202 and HLA-Cw*0801, whereas the HLA-DR0301, HLA-Cw1502 and HLA-A*0201 alleles play the role of protection against SARS infection[18]. MHC-II molecules, in case of MERS-CoV infection, such as HLA-DRB1*11:01 and HLADQB1* 02:0 represents the susceptibility characteristics of MERSCoV infection[19,20]. Besides, Mannose binding lectin (MBL) plays role to the risk of SARS-CoV infection. [20]. All these studies give us idea about the prevention, treatment & the mechanism of SARS-CoV [15].
Humoral & Cellular Immunity
Body’s humoral and cellular immunity gets stimulated due to the presence of antigen, which is being mediated by the virus specific B & T cells. Antibody profile of SARS-CoV has the same pattern of IgM & IgG production, which is similar to common acute viral infections. The SARS-specific IgE antibody disappeared by 12 weeks, whereas IgG antibody lasts for a long time, indicating that IgG antibody plays protective role [21]. These SARS-specific IgG antibodies are primarily S-specific & N-specific antibodies [22]. Latest report shows that number of CD4+ and CD8+ T cells in the peripheral blood of SARS-CoV-2 infected patients have been significantly reduced, despite having excess activation. The response in the acute phase of SARS-CoV patient depends on the decrease of CD4+ and CD8+ cells. CD4+ and CD8+ memory T cells can survive for at least 4 years in case of SARS-CoV recovered individual even without the presence of antigen. Also, it performs T cell proliferation, DTH response and IFN-Gamma production [23]. Even after 6 years of SARS-CoV infection, specific T cell memory responds to the SARS-CoV due to the presence of S peptide library in case of 14 of 23 recovered SARS patients [15,24].
Cytokine storm in COVID-19
Acute respiratory distress syndrome (ARDS) is the common immune pathological happening for SARS-CoV, SARS-CoV-2 & MERS-CoV infections [25]. One of the main mechanisms of Acute respiratory distress syndrome (ARDS) is the cytokine storm, which results in severe inflammatory response due to the presence of immune effector cells. This immune effector cells helps to release large amounts of pro-inflammatory cytokines & chemokines in case of SARS-CoV infection [26]. Attack of the cytokine storm leads to ARDS & multiple organ failure, causing death in severe cases of SARS-CoV-2 infection[15,25].
Coronavirus Immune Evasion
In order to survive, SARS-CoV & MERS-CoV induces the production of pattern recognition receptors known as PRRs by avoiding host detection of their dsRNA [27,28]. These PRR helps to recognize pathogen associated molecular pattern, known as PAMPs. Recognized by pattern recognition receptors (PRRs). SARS-CoV & MERS-CoV can induce the production of double membrane vesicle that lacks PRRs, then replicate in these vesicles. IFN-1 shows protective effect against SARS-CoV & MERS-CoV, but by activating MDA5, accessory protein 4a of MERS-CoV blocks the induction of IFN by directly interacting with the double stranded RNA [29]. ORF4a, ORF4b, ORF5, and membrane proteins of MERS-CoV also inhibits the nuclear transport of IFN regulatory factor 3 and activation of TFNBeta promoter [30]. Therefore, destroying immune evasion causes hamper in the treatment & drug development of SARS-CoV-2[15]. Black seed, scientifically known as ‘Nigella Sativa’ has been used as an effective treatment option in various kinds of diseases especially in Islamic traditional medicine. There are a lot of scientific articles which has proved earlier that Black seed has Immunomodulatory, antiviral and anti-inflammatory effects. Besides, it also protects the lung tissue. For all of these reasons, we have selected this plant for our project. Nigella seed and oil contains many kinds of ingredients. Among all the constituents, we have chosen Thymoquinone and Carvacrol which accounts (30-48%) and (6-12%) of ingredients respectively. Thymoquinone has known to exert its’ anti-inflammatory effects by inhibiting TNF-alpha. [31] There are some evidences that nutritional interventions such as vitamin and minerals can be considered as a potential treatment for COVID-19. Ascorbic acid has powerful anti inflammatory and antioxidant properties, which can be proved very effective for this disease. For this reason, we have chosen Vitamin C (Ascorbic acid) as an option. Vitamin D3 (Cholecalciferol) plays a key role in the immunity. If the immune system is strong and well functioning, we hypothesize that any type of viral infection can be prevented and cured. So, for this reason we have chosen this for our study.
Sars-Cov-2 pandemic is an emergence situation globally which has already infected millions of people and taken a lot of lives. The world needs an effective medicine for the treatment of COVID 19 caused by this novel corona virus. Invention of a new medicine is quite long time consuming and cost inefficient. There are a lot of antiviral drugs available in the market for the treatment of different viral diseases. We picked 16 of those for our molecular docking study against the main enzyme RNA dependent RNA polymerase of Sars- Cov-2 which is responsible for the viral replication in the host cells. After evaluating some performance parameters, we will recommend couple of molecules for further study in search of an inhibitor of Sars- Cov-2 for the treatment of COVID 19. Further study with already available medicines will save time and money altogether to discover a new treatment.
Materials and Methods
To find the effective drug against SARS-COV-2, 3D structure of the SARS-COV-2 RDRP (RNA Dependent RNA Polymerase) is needed. All the selected ligands structure were downloaded from the PubChem database. For this purpose, the structure of the protein is obtained from Protein Data Bank (RCSB PDB).
Data Collection
Protein Data Bank (RCSB PDB) was used for the collection of the structure of SARS-COV-2 RDRP. The structure of the SARS-COV-2 RNA Dependent RNA Polymerase (6m71) was downloaded from this database.
Validation of the Structure
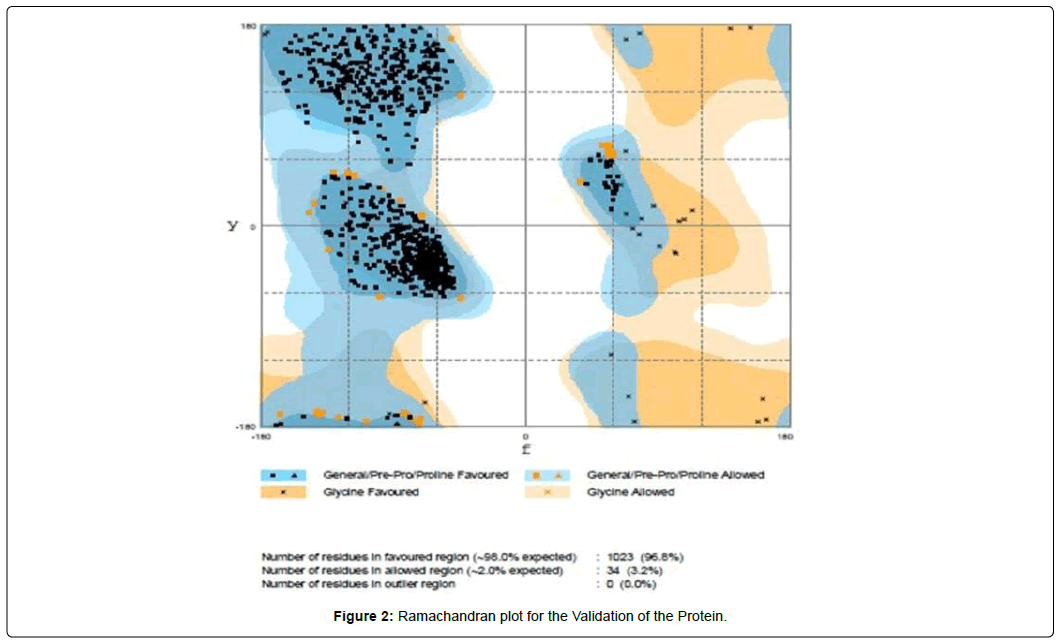
We have validated the structure by using Ramachandran plot which is available at the RAMPAGE webserver.
Pocket Formation
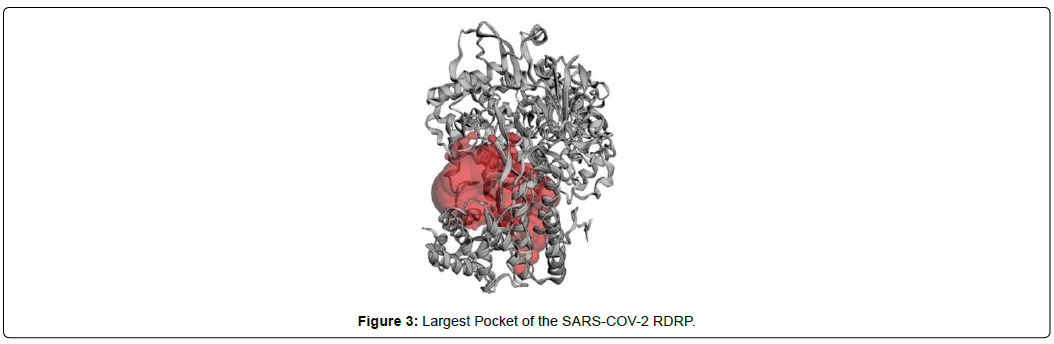
In order to find a suitable pocket, CASTp webserver was used to determine pockets and their specific dimensions. For the docking process, we selected the largest pocket of the protein as it is considered as the best binding site of a ligand according to the rule of thumb.
Molecular Docking
Molecular docking was conducted using the software Auto dock Vina which is freely available in the MGLtools website [32-37].
Result
In this section, all the results obtained from each step mentioned in the previous section are shown in the respective manner as presented in methodology.
PDB Structure of the SARS-COV-2 RNA Dependent RNA Polymerase (RDRP)
The PDB structure of the SARS-COV-2 RDRP was obtained from the Protein Data Bank (Figure 1).
Validation by Ramachandran Plot
In the Ramachandran plot, it has been shown that the number of residues in favored region was 96.8%, which is well above the standard range. So, we can easily assume that the structure is truly validated (Figure 2).
Docking
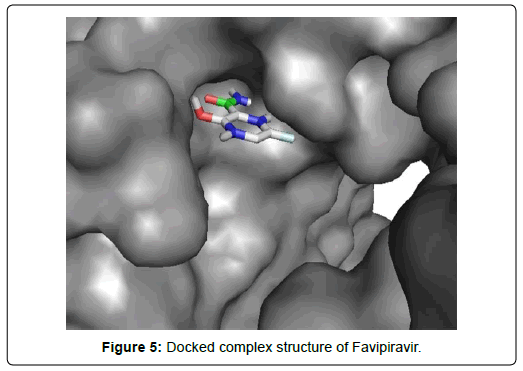
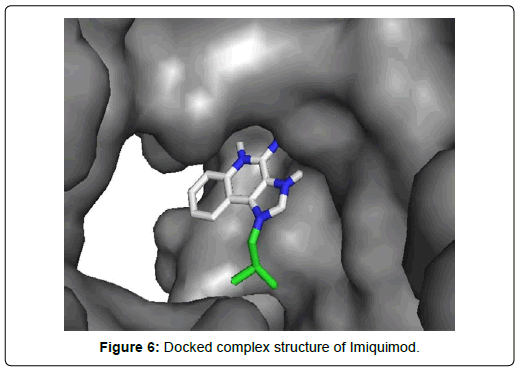
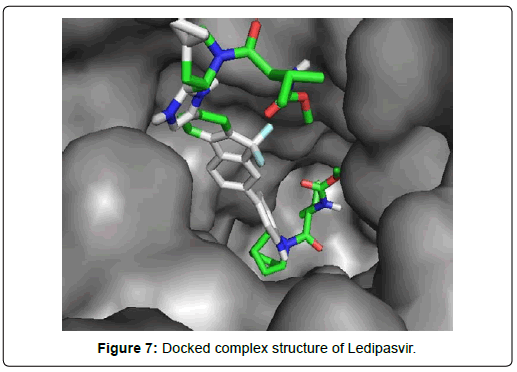
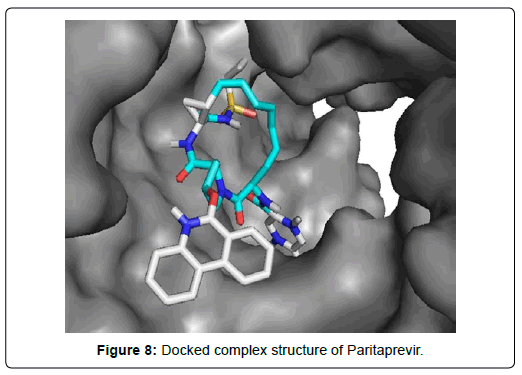
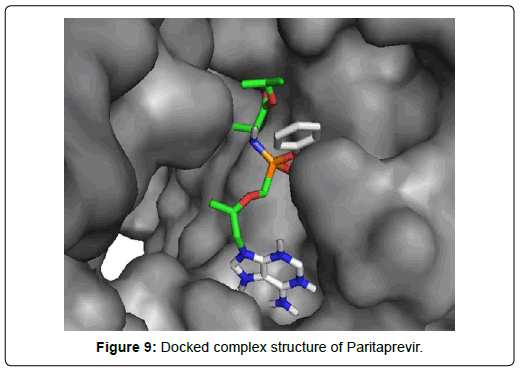
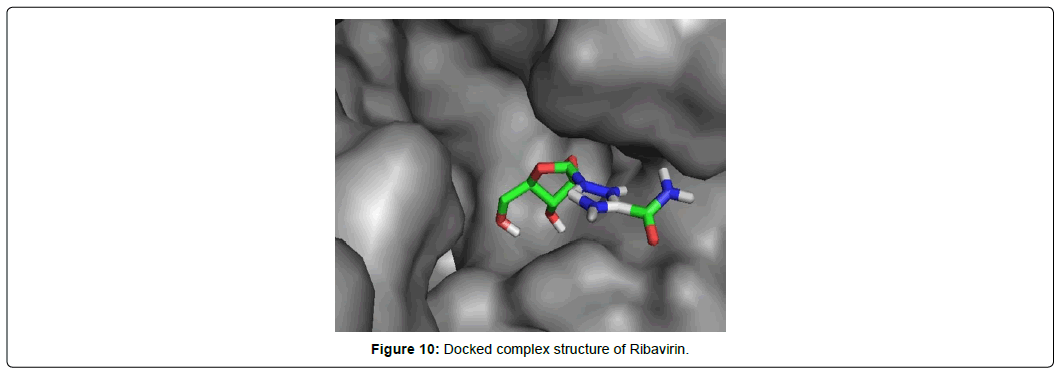
When the selected molecules were docked with the crystal structure of the SARS-COV-2 RDRP, it produced 10 conformations for each ligand (Figure 3). The docking process resulted with specific binding affinities and RMSD values of the ligands. After the docking process, we compared all the selected ligands binding affinities with each other. Finally, we discovered that Remdesivir is the best candidate against SARS-COV-2 RDRP (Figures 4-10).
Discussion
Molecular docking study was performed against RNA dependent RNA polymerase (RDRP) enzyme of Sars-Cov-2 virus with a group of 20 ligands that included 16 currently available antiviral drugs used in different viral diseases, 2 vitamins and 2 phytochemical components. We studied binding affinity using auto dock vina and analyzed amino acid interactions of ligands with the protein using discovery studio. Before putting the ligands on autodock vina, we prepared the ligands adding protons and charges using UCF Chimera. The Sars-Cov-2 RDRP protein was also evaluated by using Ramachandran plot. The parameters we considered in evaluating the docking performance are docking scores, RMSD values and amino acid interactions between ligand and protein.
Docking score: Docking scoring is used to evaluate which of the conformations of a ligand possess the best complement to the protein binding site. A well docked complex yields more negative docking score than a poorly docked complex.Docking scores between the ligands and Sars-Cov-2 RDRP virus were generated by Autodock vina.
RMSD value: Root-mean-square deviation (RMSD) can be defined as the average distance between the atoms of structures those are superimposed on one another. RMSD value is one of recognized parameters to evaluate the performance of molecular docking. The docking accuracy is primarily deemed based on RMSD value of the atoms of a ligand in the docked pose with protein from the atoms of the crystal structure. A lower RSMD value indicates that the conformation is more stable than conformations those have higher RMSD value. A RMSD value (upper bound) with less than 2.0Å is recommended. In regards docking accuracy, a RMSD value (upper bound) between 1.0-3.0 Å between a docked and X-ray pose is usually considered as a successful docking. The generation of a good RMSD value depends on the rotatable bonds in ligands. The desired RMSD values of less than 2.0 Å mainly results when there are two to six rotatable bonds present in a ligand. The more the number of rotatable bonds present the higher the RMSD value results. In cross docking experiments, when a ligand is docked with an apo structure of a protein, the main position of the ligand is usually incurred by superimposing structures of holo and apo protein. This alignment can be influenced by the differences between the poses of the holo and apo proteins and also by the group of atoms used for the superposition. Therefore, RMSD value (upper bound) may be little higher than the recommended value in cross docking experiments.
Therefore, we relaxed the cutoff RMSD value (upper bound) to 2.5Å to evaluate the performance of our docking. We considered the lowest RMSD value (upper bound) to the standard among all the poses for each and every ligand [Table 1]. Then we picked up top 10 ligands those have RMSD values (upper bound) less than 2.5Å [Table 2].
| SI | Name of ligand | Docking Score (KJ/mol) | Lowest RMSD (Upper bound) |
|---|---|---|---|
| 1 | Boceprevir | -7.4 | 2.05 |
| 2 | Dolutegravir | -7.3 | 6.2 |
| 3 | Elvitegravir | -6.5 | 2.05 |
| 4 | Entecavir | -6.3 | 2.5 |
| 5 | Favipiravir | -5.8 | 2.14 |
| 6 | Fosamprenavir | -6.2 | 5.65 |
| 7 | Imiquimod | -5.5 | 2.45 |
| 8 | Ledipasvir | -9.4 | 2.24 |
| 9 | Letermovir | -7.2 | 8.29 |
| 10 | Nelfinavir | -7.7 | 6.15 |
| 11 | Oseltamivir | -4.9 | 2.76 |
| 12 | Paritaprevir | -9.7 | 2.19 |
| 13 | Ribavirin | -5.8 | 1.93 |
| 14 | Sofosbuvir | -7.1 | 2.49 |
| 15 | Tenofovir Alafenamide | -6.6 | 2.41 |
| 16 | Tenofovir Disoproxil | -6.1 | 7.87 |
| 17 | Thymoquinone | -4.5 | 2.51 |
| 18 | Ascorbic Acid | -4.6 | 3.01 |
| 19 | Carvacrol | -4.3 | 3.51 |
| 20 | Cholecalciferol | -6 | 2.28 |
Table 1: Binding affinities (KJ/mol) of all 20 ligands with their lowest RMSD value (upper bound).
| SI | Name of ligand | Docking Score (KJ/mol) | Lowest RMSD (Upper bound) |
|---|---|---|---|
| 1 | Boceprevir | -7.4 | 2.05 |
| 2 | Elvitegravir | -6.5 | 2.05 |
| 3 | Favipiravir | -5.8 | 2.14 |
| 4 | Imiquimod | -5.5 | 2.45 |
| 5 | Ledipasvir | -9.4 | 2.24 |
| 6 | Paritaprevir | -9.7 | 2.19 |
| 7 | Ribavirin | -5.8 | 1.93 |
| 8 | Sofosbuvir | -7.1 | 2.49 |
| 9 | Tenofovir Alafenamide | -6.6 | 2.41 |
| 10 | Cholecalciferol | -6 | 2.28 |
Table 2: Binding affinities (KJ/mol) of 10 ligands sorted out in first screening based on the RMSD values (upper bound) less than 2.5.
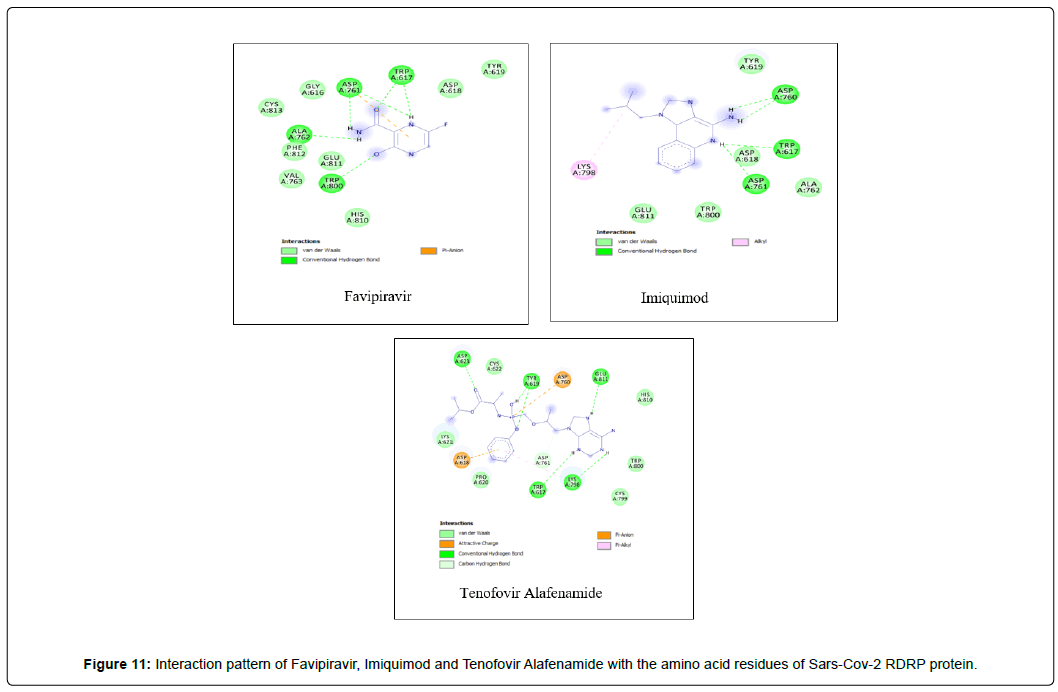
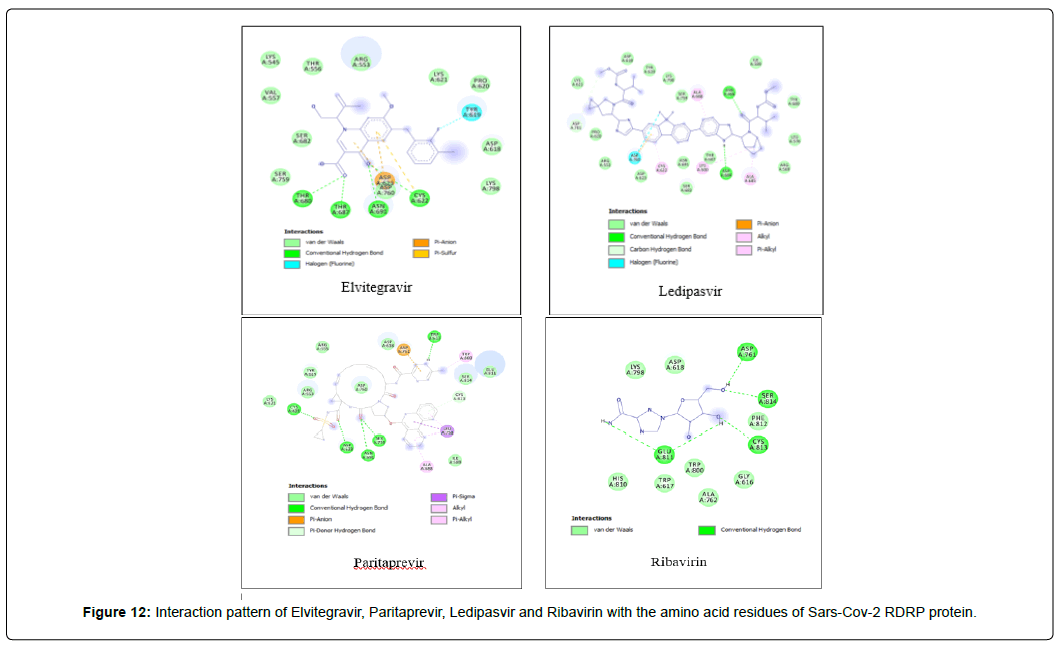
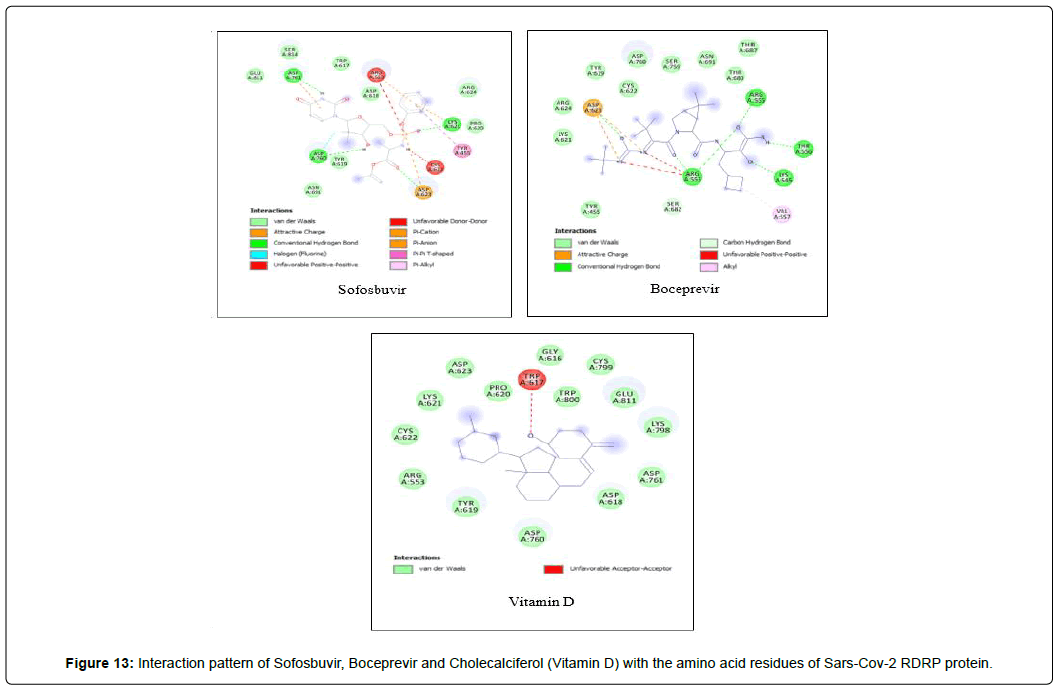
Amino acid interactions: Analysis of amino acid interaction is an important parameter in evaluating the performance of molecular docking. A good number of hydrophobic interactions e.g Van der waals and hydrogen bond interactions stabilize the complex between protein and a ligand.The presence of hydrogen bonds in proteinligand interaction plays important role to yield a good binding affinity. We studied the amino acid interaction in our molecular docking using discovery studio. We again sorted out 7 ligands from Table 2 depending upon amino acid interaction patterns [Table 3].Among all 20 ligands we sorted out Boceprevir, Elvitegravir, Favipiravir, Imiquimod, Ledipasvir, Paritaprevir, Ribavirin, Sofosbuvir, Tenofovir Alafenamide and Cholecalciferol in first screening because these ligands possessed RMSD value less than 2.5 Å [Table 2]. Amongst the selected 11 ligands, Paritaprevir and Ledipasvir yielded two best binding affinities of -9.7 and 9.4 KJ/mol with RMSD value of 2.19 and 2.24 Å respectively. Boceprevir presented a docking score of -7.4 KJ/mol along with a good RMSD value of 2.05 Å. Elvitegravir demonstrated similar kind of result like Boceprevir with a docking score of -6.5 KJ/mol and RMSD value of 2.05 Å. Ribavirin possessed the best RMSD value of 1.93 Å among the selected 11 ligands in first screening with a docking score of -5.8 KJ/mol. Besides, Cholecalciferol which is also known as Vitamin D3 exhibited a docking score of with -6 KJ/mol along with a RMSD value of -2.28 Å. Then we performed second screening among the 10 ligands sorted from first screening based on the amino acid interactions between protein and the ligands. We picked up 7 ligands those exhibited a good number of interactions including Van der waals and hydrogen bond interactions [Figure 11 and 12]. The remaining ligands were eliminated because of either very limited number of interactions with the protein or unfavorable bumps in the interactions. Unfavorable bumps can affect the stability of the complex because these bonds may cause force repulsion between two molecules and an atom.We screened out Boceprevir, Sofosbuvir and Cholecalciferol because these molecules showed one or more unfavorable bumps in the amino acid interactions [Figure 13].
| SI | Name of ligand | Docking Score (KJ/mol) | RMSD (Upper bound) |
|---|---|---|---|
| 1 | Elvitegravir | -6.5 | 2.05 |
| 2 | Favipiravir | -5.8 | 2.14 |
| 3 | Imiquimod | -5.5 | 2.45 |
| 4 | Ledipasvir | -9.4 | 2.24 |
| 5 | Paritaprevir | -9.7 | 2.19 |
| 6 | Ribavirin | -5.8 | 1.93 |
| 7 | Tenofovir Alafenamide | -6.6 | 2.41 |
Table 3: Binding affinities (KJ/mol) of final 7 molecules with their RMSD values (upper bound) sorted out in second screening based on the amino acid interaction pattern. Here, all 7 ligands don’t have any unfavorable bumps.
Favipiravir displayed total 12 amino acid interactions through two different types of bonds which included 8 van der waals and 4 conventional hydrogen bonds. Imiquimod formed total 9 residual three different types of interactions including 5 van der waals, 3 hydrogen bonds and 1 alkyl bond. Tenofovir Alafenamide displayed total 14 amino acid interactions consisting 6 van der waals, 5 hydrogen bonds, 1 carbon-hydrogen bond, 1 attractive charge and 1 pi-anion bond.
Elvitegravir formed total 17 residual interactions with four different types of interactions (11 van der waals bonds, 4 conventional hydrogen bonds, 1 fluoride interaction and 1 pi-anion bond), Paritaprevir was involved with 19 residual interactions possessing six different types interactions including 8 van der waals bonds, 5 hydrogen bonds, 2 pi-donor hydrogen bonds, 1 alkyl bond, 1 pi-alkyl bond and 1 pi-anion bond. Ledipasvir exhibited total 23 interactions forming six different interactions (15 van der waals bonds, 2 conventional hydrogen bonds, 1 halogen (fluoride) bonds, 1 carbon-hydrogen bond, 2 alkyl bonds and 2 pi-alkyl bonds. Ribavirin interacted with total 12 amino acid residues in two bonding types comprising 8 van der waals and 4 conventional hydrogen bonds. Figure 13: Interaction pattern of Sofosbuvir, Boceprevir and Cholecalciferol (Vitamin D) with the amino acid residues of Sars-Cov-2 RDRP protein. All three ligands displayed unfavorable bumps (red dotted lines).
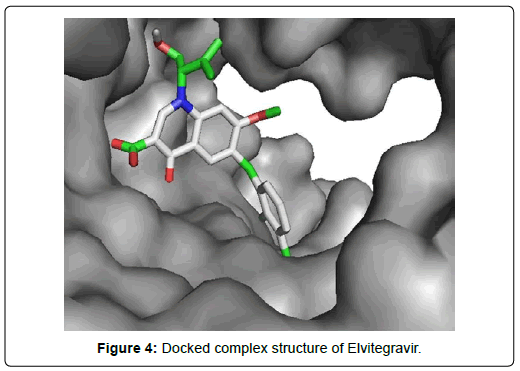
Among the final 7 ligands those are sorted out in second screening, Elvitegravir demonstrated total 17 residual interactions with four different types of interactions with the protein. The interactions include 11 van der waals bonds with VAL A:557, THR A:556, LYS A:621, ASP A:618, SER A : 759, LYS A:545, ARG A : 553, PRO A : 620, LYS A : 798, SER A: 682, ASP A :760 residues, 4 conventional hydrogen bonds with THR A: 680, ASN A: 691,THR A: 687, CYS A:622 residues, 1 fluoride interaction with TYR A:619 and 1 pi-anion bond with ASP A: 623 residue. The only halogen (fluoride) bond of Elvitegravir with the protein will play important role to stabilize the complex. This non-covalent interaction may also improve biological activity substantially [34] Thus, Elvitegravir can be an effective inhibitor of Sars-Cov-2 RDRP based on the analysis of its docking score (-6.5 KJ/mol), rmsd value (2.05 Å) and interaction pattern with the amino acids.
Paritaprevir was involved with 19 residual interactions possessing six different types interactions including 8 van der waals bonds with ARG A: 555, ARG A:553, ASP A: 760, SER A:814, ASP A: 618, TYR A:619, LYS A: 621, GLU A : 811 residues, 5 hydrogen bonds with CYS A: 622, ASN A:691, TRP A: 617, ASP A:623, SER A:759, 2 pidonor hydrogen bonds with CYS A:813 and ILE A:589 residues, 1 alkyl bond with ALA A:688, 1 pi-alkyl bond with TRP A: 800 and 1 pi-anion bond with ASP A:761 amino acid residue. The abundance of different interactions of Paritaprevir comprising a good number of hydrogen bonds and other hydrophobic bonds, a good docking score (-9.7 KJ/mol) and rmsd value (2.19 Å) makes the ligand a competent candidate for being a Sars-Cov-2 RDRP inhibitor.
Ledipasvir exhibited the highest total 23 interactions with amino acid residues of the protein among all 8 ligands selected in second screening. It also made six different interactions with the protein. The interactions included 15 van der waals bonds with LYS A: 621 , ARG A: 553, ASN A: 691, THR A: 687 , LEU A: 576, ILE A: 598, LYS A: 798, ASP A: 618, PRO A: 620, ASP A: 623, SER A: 682, ARG A:569, TYR A:689, SER A: 759, TYR A: 619 residues, 2 conventional hydrogen bonds with ASP A:684, ASN A:496 residues, 1 halogen (fluoride) bonds with ASP A: 760, 1 carbon-hydrogen bond with ASP A : 761, 2 alkyl bonds with CYS A:622 and LYS A:500 and 2 pi-alkyl bonds with ALA A:685 and ALA A: 688 residues. The number of hydrogen bonds is lesser in Ledipasvir than in Paritaprevir and Elvitegravir. Nevertheless, a good number of van der waals interactions, couple of hydrogen bonds, a halogen bond along with some other interactions, a docking score of -9.4 KJ/mol with rmsd value of 2.24 Å makes Ledipasvir also a good candidate for further analysis to be an inhibitor of Sars-Cov-2 virus RDRP.
Ribavirin having the best rmsd value of 1.93 Å among the final 8 ligands demonstrated total two types of interactions included hydrophobic van der Waals and conventional hydrogen bonds. It interacted with total 12 amino acid residues comprising 8 van der waals with LYS A: 798, HIS A: 810, TRP A: 800, GLY A: 616, ASP A: 618, TRP A: 617, ALA A: 762, PHE A: 812 and 4 conventional hydrogen bonds with ASP A: 761, SER A: 814, CYS A: 813, GLU A: 811 residues. The lowest RSMD value Ribavirin gave it the most stable conformation among all ligands.
Favipiravir displayed total 12 amino acid interactions including 8 van der waals and 4 conventional hydrogen bonds like Ribavirin. Favipiravir interacted CYS A: 813, PHE A: 812, VAL A: 763, ASP A: 618, GLY A: 616, GLU A: 811, HIS A: 810, TYR A: 619 residues with van der waals bonds and interacted ALA A: 762, TRP A: 617, TRP A: 800, ASP A: 761 with hydrogen bonds.
Imiquimod and Tenofovir Alafenamide both have rmsd values (2.45 and 2.41 Å) little less than 2.5 Å along with docking scores of -5.5 and 6.6 KJ/mol respectively. Imiquimod exhibited total 9 residual three different types of interactions including 5 Van der waals with GLU A: 811, ALA A: 762, TYR A: 619, TRP A: 800, ASP A : 618, 3 hydrogen bonds with ASP A: 761, TRP A:617, ASP A: 760 and 1 alkyl bond with LYS A: 798 residue. It possessed the least number of interactions among the final 7 molecules. Tenofovir Alafenamide displayed total 14 amino acid interactions consisting 6 Van der waals with LYS A: 621, CYS A:799, HIS A: 810, PRO A:620, TRP A: 800, CYS A:622, 5 hydrogen bonds with ASP A: 623, GLU A: 811, TRP A:617, TYR A:619, LYS A:798, 1 carbon-hydrogen bond with ASP A: 761, 1 attractive charge with ASP A:618 and 1 pi-anion bond with ASP A: 760.
Out of 20 ligands, after two successive screenings we selected seven ligands those fulfilled the criteria of the parameters we set up to evaluate the overall docking performance. All seven ligands (Paritaprevir, Elvitegravir, Ledipasvir, Ribavirin, Favipiravir, imiquimod and Tenofovir Alafenamide) are currently available antiviral medicines used for the treatment of different viral infections. There were two vitamins (Ascorbic acid and Cholecalciferol) and two phytochemical components (Thymoquinone and Carvacrol) in the primary list, all of them were eliminated in the screenings either because of having high RMSD value or for possessing unfavorable bumps in the interactions or for having limited number of interactions.
Conclusion
In pursuit of finding a Sars-Cov-2 RNA dependent RNA polymerase enzyme inhibitor for the treatment of COVID 19 (Corona Virus Disease 2019) we performed a molecular docking study with 20 ligands among of them 16 ligands are currently available antiviral medicines, 2 are vitamins and 2 are phytochemical components. After analyzing the binding affinities, RMSD values (upper bound) and amino acid interactions of the ligands we short listed seven molecules those have passed all the parameter of performance evaluation. Amongst, we recommend five molecules (Paritaprevir, Elvitegravir, Ledipasvir, Ribavirin and Favipiravir) for further research in search of Sars-Cov-2 RDRP inhibitor(s) for the treatment of COVID 19.
References
- Cui J, Li F, Li Shi Z (2018) Origin and evolution of pathogenic coronaviruses. Nature Reviews Microbiology 17:181–192.
- Huang C, Wang Y, Li X (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 395: 497-506.
- Lu H, Stratton W C, Tang W Y(2020) Outbreak of pneumonia of unknown etiology in Wuhan, China: The mystery and the miracle. Journal of Medical Virology 92: 401-402.
- Shereen A, Khan M, Kazmi S, Bashir A, Siddique N et al (2020) COVID-19 infection: origin, transmission and characteristics of human coro-naviruses. Journal of advance research 24: 91-98.
- Wang N, Shi X, Jiang L, Zhang S, Wang D, et al (2013) Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Research 23:986-993.
- Glowacka I, Bertram S, Muller M. A, Allen P, Soilleux E et al (2011) Evidence that TMPRSS2 Activates the Severe Acute Respiratory Syndrome Coronavirus Spike Protein for Membrane Fusion and Reduces Viral Control by the Humoral Immune Response. Journal of Virology 85:4122–4134.
- Bertram S, Glowacka I, Muller M. A, Lavender H, Gnirss K et al (2011) Cleavage and Activation of the Severe Acute Respiratory Syndrome Coronavirus Spike Protein by Human Airway Trypsin-Like Protease. Journal of Virology 85: 13363–13372.
- Fung S To, Liu X Ding, (2014) Coronavirus infection, ER stress, apoptosis and innate immunity. frontiers in Microbiology 5: 1-13.
- Lu Y, Lu X, Denison R M (1995) Identification and Characterization of a Serine-Like Proteinase of the Murine Coronavirus MHV-A59. Journal of Virology 69: 3554–3559.
- Sahin AR, Erdogan A, Mutlu Agaoglu P, Dineri Y, Cakirci AY, et al (2020) 2019 Novel Corona virus (COVID 19) Outbreak : A Review of the Current Literature. Eurasian journal of Medicine and Oncology 4: 1-7.
- Wit DM, Doremalen VN, Falzarano D, Munster JV(2016) SARS and MERS: recent insights into emerging coronaviruses. Nature Reviews Microbiology 14: 523–534.
- Wu F, Zhao S, Yu B, Chen M Y, Wang W et al, (2020) A new coronavirus associated with human respiratory disease in China, Nature, 579:265–269.
- Millet KJ, Whittaker R G (2014) Host cell entry of Middle East respiratory syndrome coronavirus after two-step, furin-mediated activation of the spike protein. PNAS 111:15214-15219.
- Perlman S, Netland J, (2009) Coronaviruses post-SARS: update on replication and pathogenesis. Nature Reviews Microbiology 7: 439-450.
- Li X, Geng M, Peng Y, Meng L, Lu S, et al (2020) Molecular immune pathogenesis and diagnosis of COVID-19. Journal of Pharmaceutical Analysis 10: 102-108.
- Wit ED, Doremalen NV, Falzarano D, Munster VJ (2016) SARS and MERS: recent insights into emerging coronaviruses. Nature Reviews Microbiology 14:523–534.
- Liu J, Wu P, Gao F, Qi J, Kawana-Tachikawa A, et al (2010) Novel Immunodominant Peptide Presentation Strategy: A Featured HLA-A*2402-Restricted Cytotoxic T-Lymphocyte Epitope Stabilized by Intrachain Hydrogen Bonds from Severe Acute Respiratory Syndrome Coronavirus Nucleocapsid Protein. Journal of Virology 84: 11849–11857.
- Wang SF, Chen KH, Chen M, Li WY, Chen YJ, et al (2011) Human-Leukocyte Antigen Class I Cw 1502 and Class II DR 0301 Genotypes Are Associated with Resistance to Severe Acute Respiratory Syndrome (SARS) Infection. Viral immunology 25:421-426.
- Hajeer AH, Balkhy H, Johani S, Yousef MZ, Arabi Y (2016) Association of human leukocyte antigen class II alleles with severe Middle East respiratory syndrome-coronavirus infection. Annals of theoretic medicines 11: 211-213.
- Tu X, Chong W. P, Zhai Y, Zhang H, Zhang F (2015) Functional polymorphisms of the CCL2 and MBL genes cumulatively increase susceptibility to severe acute respiratory syndrome coronavirus infection. Journal of Infection 71: 101-109.
- Li G, Chen X, Xu A (2003) Profile of Specific Antibodies to the SARS-Associated Coronavirus. New England Journal of Medicine 349: 508–509.
- Li X, Geng M, Peng Y, Meng L, Lu S (2020) Molecular immune pathogenesis and diagnosis of COVID-19. Journal of Pharmaceutical Analysis 10: 102-108.
- Fan YY, Huang ZT, Li L, Wu MH, Yu T, et al (2009) Characterization of SARS-CoV-specific memory T cells from recovered individuals 4 years after infection. Archives of Virology 154:1093–1099.
- Tang F, Quan Y, Xin ZT, Wrammert J, Ma MJ, et al (2015) Lack of Peripheral Memory B Cell Responses in Recovered Patients with Severe Acute Respiratory Syndrome: A Six-Year Follow-Up Study. The journal of immunology 186:7264-7268.
- Xu Z, Shi L, Wang Y, Zhang J, Huang L, et al (2020) Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet respiratory medicine 8: 420-422.
- Huang C, Wang Y, Li X, Ren L, Zhao J, et al (2020) Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. The Lancet 395: 497–506.
- Snijder EJ, Van der Meer Y, Zevenhoven-Dobbe J, Onderwater JJ., Van der Meulen J, et al (2006) Ultrastructure and Origin of Membrane Vesicles Associated with the Severe Acute Respiratory Syndrome Coronavirus Replication Complex. Journal of Virology 80: 5927–5940.
- Niemeyer D, Zillinger T, Muth D, Bielecki F, Horvath G, et al (2013) Middle East Respiratory Syndrome Coronavirus Accessory Protein 4a Is a Type I Interferon Antagonist. Journal of Virology 87:12489–12495.
- Yang Y, Zhang L, Geng H, Deng Y, Huang B, et al (2013) The structural and accessory proteins M, ORF 4a, ORF 4b, and ORF 5 of Middle East respiratory syndrome coronavirus (MERS-CoV) are potent interferon antagonists. Protein & Cell 4: 951–961.
- Umar S , Hedaya O, Sing AK, Ahmed S (2015) Thymoquinone inhibits TNF-α-induced inflammation and cell adhesion in rheumatoid arthritis synovial fibroblasts by ASK1 regulation. Toxicology and Applied Pharmacology 287: 299-305.
- Warren GL, Andrews CW, Capelli AM, Clarke B, LaLonde J, et al (2006) A Critical Assessment of Docking Programs and Scoring Functions. Journal of Medicinal Chemistry 49: 5912-5931.
- Xiao W, Wang D, Shen Z, Li S, Li H (2018) Multi-Body Interactions in Molecular Docking Program Devised with Key Water Molecules in Protein Binding Sites. Molecules 23: 2321.
- Kurczab R, Kucwaj-Brysz K, Sliwa P (2019) The Significance of Halogen Bonding in Ligand– Receptor Interactions: The Lesson Learned from Molecular Dynamic Simulations of the D4 Receptor. Journal of medicinal chemistry 25: 91.
- Erickson JA, Alaie MJ, Robertson DH, Lewis RA, Vieth M (2004) Lessons in Molecular Recognition: The Effects of Ligand and Protein Flexibility on Molecular Docking Accuracy. Molecules 47: 45-55.
- Ravindranath PA, Forli S, Goodsell DS, Olson AJ, Sanner MF (2015) AutoDockFR: Advances in Protein-Ligand Docking with Explicitly Specified Binding Site Flexibility. Plos Computational Biology 11:28.
- Vargas JAR, Lopez AG, Froeyen M, Piñol MC, Froeyen M (2018) Molecular docking study on the interaction between 2-substituted-4,5-difuryl Imidazoles with different Protein Target for antileishmanial activity. Journal of Applied Pharmaceutical Science 8: 014-022.
- Dhorajiwala TM, Halder ST, Samant L (2019) Comparative In Silico Molecular Docking Analysis of L-Threonine-3-Dehydrogenase, a Protein Target Against African Trypanosomiasis Using Selected Phytochemicals. Journal of Applied Biotechnology Reports 6: 101-108.