Research Article, J Appl Bioinforma Comput Biol Vol: 6 Issue: 2
Marker Association Analysis with Three Agronomic Traits in Hard Winter Wheat Lines under Diverse Environments
Yi Xu1, Guihua Bai2, Robert A Graybosch3, Yajun Wu4 and Jixiang Wu1*
1Department of Agronomy, Horticulture, and Plant Science, South Dakota State University, South Dakota, USA
2U.S. Department of Agriculture-Agricultural Research Service, Hard Winter Wheat Genetics Research Unit, Kansas, USA
3U.S. Department of Agriculture-Agricultural Research Service, 137 Keim Hall, University of Nebraska, Nebraska, USA
4Department of Biology and Microbiology, South Dakota State University, South Dakota, USA
5Department of Mathematics and Statistics, South Dakota State University, South Dakota, USA
*Corresponding Author : Jixiang Wu
Department of Agronomy, Horticulture, and Plant Science, Box 2140C, South Dakota State University, Brookings, SD 57007, USA
Tel: 605-688-5947
E-mail: jixiang.wu@sdstate.edu
Received: March 19, 2017 Accepted: July 13, 2017 Published: July 19, 2017
Citation: Xu Y, Bai G, Graybosch RA, Wu Y, Wu J (2017) Marker Association Analysis with Three Agronomic Traits in Hard Winter Wheat Lines under Diverse Environments. J Appl Bioinforma Comput Biol 6:2. doi: 10.4172/2329-9533.1000136
Abstract
Genetic performance for a quantitative trait is often controlled by gene-by-gene and genotype-by-environment (GE) interaction effects. As an important component, a GE interaction effect is highly related to crop stability. As genetic association mapping has been widely used to determine markers associated with traits of interest, stability analysis based on genetic markers/genes of importance could help develop widely adapted or specifically adapted cultivars. With linear mixed model approach, in this study we analyzed a wheat data set from the USDA Hard Winter Wheat Regional Nursery Program in 2010. The data included 32 genetic markers and 48 genotypes with three important agronomic traits: grain yield, plant height, and heading date, which were measured under multi-environments. Our results showed that four important DNA markers: Rht2, PPO18NED, Lr34JagTM, and Waxy-A1-AFC-AR2FAM, were significantly associated with all these three traits. Among these, Rht2 contributed 50.34%, 78.65%, and 53.90% of the phenotypic variation for grain yield, plant height, and heading date, respectively. Compared to their main effects, however, genotype-by-environment interaction effects were less important under these diverse environments.
Keywords: Gene expression; Winter wheat; Genetic marker; Agronomic trait.
Introduction
Wheat, as an important source of protein, vitamins and minerals [1], is a major consumed food crop around the world. The world production of wheat in 2012 was 670 million metric tons, making it the second most-produced cereal after rice (719 million metric tons) [2]. Wheat feeds 4.5 billion people in 95 developing countries [3]. As the population continues to increase, genetic improvement in wheat yield, quality, and resistance is more urgent to meet such a great need.
Association mapping has been widely used in detecting genetic markers associated with traits of importance that can be used for crop/animal improvement [4-9]. Various useful statistical methods and computing tools have been developed to meet such a great need for association mapping studies [10-21]. These methods have been applied to various plant species, including wheat [22-27], barley [28-31], rice [32-35] soybean [36-38] and cotton [39,40]. However, many association mapping studies were focused on studies under single or a few environments. For example, the numbers of environments used for many wheat association mapping were normally fewer than five [41,42] yet only a few studies used a little large number of environments for association mapping in wheat [43].
The Hard Winter Wheat Regional Nursery (HWWRN) Program, coordinated by U.S. Department of Agriculture - Agricultural Research Service (USDA-ARS), aims to evaluate various advanced breeding lines and commercial winter wheat cultivars in multi-state environments. More importantly, these winter wheat genotypes have been genotyped with various DNA markers with potentially known gene functions. For example, the major dwarfing genes like Rht1 andRht2, which could reduce plant height by reducing the response to gibberellin with pleiotropic effects on grain number and yield [43,44] were used to genotype 48 winter wheat lines in 2010. Various DNA markers linked to important genes in hard winter wheat were reviewed in a recent publication [45]. Such resources could provide a great opportunity for both yield stability analysis for each winter wheat cultivar, but also for determination of DNA marker associations with traits of interest under a wide range of growing environments. Appropriate genetic data analysis should provide useful genetic information to determine appropriate winter wheat cultivars. With such wheat data under multi-environments, various genetic models can be employed to reveal rich genetic information that can be used for winter wheat line selection.
In this study, we focused on analysis of HWWRN data collected by 20 institutes in 2010. The data included three important agronomic traits: grain yield, plant height, and heading date measured under diverse environments and 32 genetic markers for 48 winter wheat cultivars. First we used a one-way ANOVA model to determine each of these DNA markers associated with traits of interest under different environments. Secondly, we applied a linear mixed model approach to investigate the contributions of several these markers associated with these three agronomic traits across environments. The main objective of this study was to determine the stability of these gene expressions under various environments and provide information to select superior genotypes with stable performance.
Materials and Methods
Materials and experiments
The nursery program includes the Southern Regional Performance Nursery (SRPN), the Northern Regional Performance Nursery (NRPN) and the Regional Germplasm Observation Nursery (RGON). The winter wheat phenotypic and genotypic data were downloaded from the Hard Winter Wheat Regional Nursery Program of U.S. Department of Agriculture, which conducted by 20 institutions in 2010. We only used the phenotypic and genotypic data from SRPN in this study. Three agronomic traits, grain yield, plant height, and heading date, were available in 30, 18 and 13 environments (Table 1) and were used in this study. Forty-eight winter wheat lines were screened with 38 genes/markers. Six markers were removed due to due their monomorphism and nine genotypes were removed from the data set due to missing markers. Therefore, the data used in this study contained 39 genotypes and 32 DNA polymorphic genes markers (10 dominant and 22 co-dominants) (Table 2).
| Environment | Abbreviation | Grain yield | Plant height | Heading date |
|---|---|---|---|---|
| Clovis, NM dryland | E1 | X† | X | X |
| Clovis, NM irr. | E2 | X | X | X |
| Farmington, NM irr. | E3 | X | X | X |
| Bushland, TX dryland | E4 | X | X | X |
| Bushland, TX irr. | E5 | X | X | X |
| Chillicothe, TX | E6 | X | ||
| Prosper, TX | E7 | X | ||
| Stillwater, OK | E8 | X | ||
| Goodwell, OK irr. | E9 | X | ||
| Lahoma, OK | E10 | X | ||
| Granite, OK | E11 | X | ||
| Akron, CO | E12 | X | X | X |
| Burlington, CO | E13 | X | ||
| Fort Collins, CO irr. | E14 | X | X | X |
| Washington, CO | E15 | X | X | |
| Hays, KS | E16 | X | X | X |
| Hutchinson, KS | E17 | X | ||
| Salina, KS | E18 | X | ||
| Colby, KS | E19 | X | X | X |
| Garden City, KS | E20 | X | X | X |
| Wichita, KS | E21 | X | X | X |
| Winfield, KS | E22 | X | ||
| Lincoln, NE | E23 | X | X | X |
| Clay Center, NE | E24 | X | X | |
| North Platte, NE | E25 | X | X | |
| Sidney, NE | E26 | X | X | |
| Alliance, NE | E27 | X | X | |
| Brookings, SD | E28 | X | ||
| Dakota Lakes, SD | E29 | X | ||
| Pine Bluffs, WY | E30 | X | X | X |
Table 1: Winter wheat agronomic traits among different environments in the Hard Winter Wheat Southern Regional Performance Nursery Program of U.S. Department of Agriculture in 2010.
| DNA Marker | Marker Number | Type |
|---|---|---|
| WMC0331NED | 1 | Co-dominant |
| Rht1 | 2 | Dominant |
| Rht2 | 3 | Dominant |
| GWM0261NED | 4 | Co-dominant |
| SNP8-FHB | 5 | Co-dominant |
| UMN10VIC | 6 | Co-dominant |
| Lr19-130NED | 7 | Dominant |
| Lr19-DomNED | 8 | Dominant |
| csLV34-Lr34FAM | 9 | Co-dominant |
| Lr34TM | 10 | Co-dominant |
| Lr34JagTM | 11 | Co-dominant |
| Lr34 | 12 | Co-dominant |
| VentriupLn2PET | 13 | Dominant |
| Sr2-STM559TGAGNED | 14 | Co-dominant |
| Sr2-X3B028F08PET | 15 | Dominant |
| Sr24#50FAM | 16 | Dominant |
| Sr25-BF145935VIC | 17 | Co-dominant |
| PPD-D1,R1,R2VIC | 18 | Co-dominant |
| BAR0012FAM | 19 | Co-dominant |
| BAR0170VIC | 20 | Co-dominant |
| CDO708FAM | 21 | Co-dominant |
| VRN-A1-SNPF | 22 | Co-dominant |
| VRN-D3-F6R8NED | 23 | Co-dominant |
| TSM0120FAM | 24 | Dominant |
| UMN19(GluA1)NED | 25 | Co-dominant |
| BxMARFAM | 26 | Co-dominant |
| UMN25(GluD1)NED | 27 | Co-dominant |
| UMN26(GluD1)PET | 28 | Co-dominant |
| PPO18NED | 29 | Co-dominant |
| PPO29NED | 30 | Dominant |
| Waxy-A1-AFC-AR2FAM | 31 | Co-dominant |
| GWM0469VIC | 32 | Dominant |
Table 2: DNA marker information.
Genetic models
Due to the winter wheat data structure, several different models such as stability analysis and gene expression analysis under different environments can be used for analyzing multi-environmental data. In this study, we emphasized two genetic models. The first genetic model is described as follows.
For a particular marker, there could be all homozygous and heterozygous combinations like AA, BB and or AB among 48 cultivars. Therefore, the model 1 actually is a one-way ANOVA model for each marker, where g could be different for different markers:
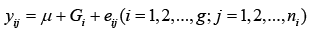 (1)
(1)
where yij is the phenotypic value for the marker with genotype i; ni is the total number of for genotype i; and 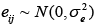 is a random error.
is a random error.
On the other hand, gene expression for each of these traits under these environments could be controlled by genotype-by-environment (GE) interaction. Investigation of GE interactions at DNA marker level is helpful to select cultivars adapted to diverse environments with specific markers. Therefore, we also analyzed each marker including its GE interactions with the following linear mixed model.
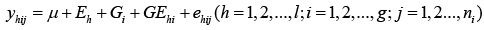 (2)
(2)
Where Eh is an environmental effect; Gi is a genotypic effect for a marker and GEhi is a GE effect for a marker and environment interaction; and ehij is a random error.
Statistical methods
With model (1), an analysis of variance (ANOVA) method was employed and adjusted coefficient of determination (R2) was estimated for each DNA marker under each environment. For model (2), a minimum norm quadratic unbiased estimation (MINQUE) was applied to estimate all variance components with 10-fold jackknife technique applied [46-48]. All data analyses were implemented by R computer language (Version 3.0.1) [49] and with an R package “minque’ [46].
Results and Discussion
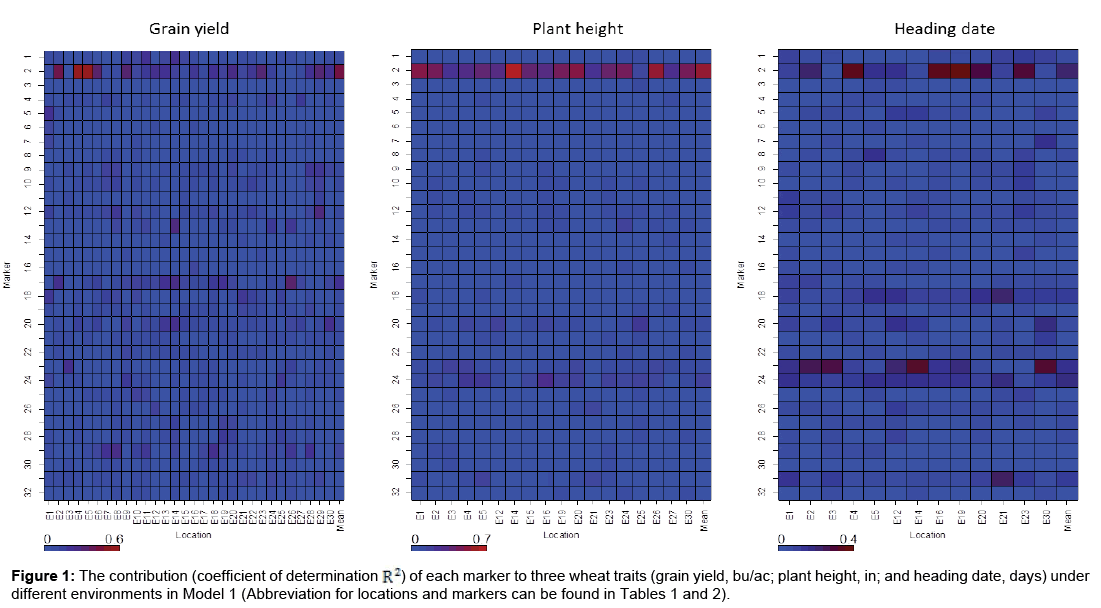
First, each marker under each environment was analyzed separately with model (1). The corresponding contribution (represented by an adjusted coefficient of determination, defined as R2) to the phenotypic variation for each of three agronomic traits was estimated. The results are provided in Figure 1.
DNA marker 2 (Rht1) made consistently higher contribution to three wheat traits than the other markers (Figure 1), indicating that Rht1 was highly associated with wheat traits grain yield, plant height, and heading date under these environments. The contribution of from this DNA marker to grain yield ranged considerably from 0.88 to 55.02% among 32 environments. As for plant height, the range of contribution was 14.81 to 71.24% among 18 environments, while for heading date, the contribution ranged from 1.05 to 38.10% among 13 environments. Rht1 was widely reported as a DNA marker related to plant anatomy and morphology, especially plant height [44,50]. The marker, VRN-D3-F6R8NED, is related to a vernalization gene and has an impact on development at stem elongation, heading date, and physiological maturity [51].
DNA marker 23, VRN-D3-F6R8NED, was highly associated with heading date (Figure 1). The contribution for VRN-D3-F6R8NED ranged from 2.18 to 32.24% for heading date among different environments. The wide ranges of genetic association with these three agronomic traits for these markers suggested that the expression of these markers under different environments varied and utilization of these markers should be environment-specific.
Single marker association analysis with three traits across environments
Each of these 32 markers was analyzed subject to Model 2 to investigate the GE interaction effects. The variance components for genotypic and GE interaction effects expressed as proportional variance components were estimated by a MINQUE approach and results are summarized in Table 3. Most markers showed small or no significant contributions to these three agronomic traits. Genotypic effects for marker Rht1 accounted for 50.34, 78.65 and 53.9% of total variations for grain yield, plant height and heading date, respectively. Genotypic effects for markers Lr34JagTM and Lr34 showed significant contributions to plant height (44.5% and 27.0%, respectively). Genotypic effects for PPO18NED had a major contribution (46.9%) to grain yield while no significant contributions to the other two traits. Genotypic effects for TSM0120FAM contributed 18.8% and 28.9% of total variations to plant height and heading date, respectively. Waxy- A1-AFC-AR2FAM contributed 13.5 and 43.0% to plant height and heading date with genotypic effects. On summary, four markers, seven markers, and nine markers made greater than 10% contribution to grain yield, plant height, and heading date, respectively. Among these markers with major contributions to these three traits, marker Rht1 showed major pleiotropic effects on all three traits. The results were in agreement with other studies [50,52,53].
Compared to genotypic effects, GE effects for these markers made small or insignificant contributions to the three traits. Among these markers only markers Rht1 and GWM0261NED had significant GE effects for grain yield (Table 3), suggesting that yield stability for these winter wheat cultivars were not related with genetic expressions of these markers and other markers or genes may be responsible for yield stability for these cultivars.
| Grain yield | Plant height | Heading date | ||||
|---|---|---|---|---|---|---|
| Marker# | VG/VP | VGE/VP | VG/VP | VGE/VP | VG/VP | VGE/VP |
| 1 | 0.020* | 0.000 | 0.005 | 0.000 | 0.044* | 0.000 |
| 2 | 0.503** | 0.147** | 0.787** | 0.027 | 0.539** | 0.069 |
| 3 | 0.059** | 0.000 | 0.020 | 0.000 | 0.077 | 0.000 |
| 4 | 0.051** | 0.081* | 0.019 | 0.000 | 0.084* | 0.000 |
| 5 | 0.011 | 0.000 | 0.000 | 0.000 | 0.084* | 0.000 |
| 6 | 0.008 | 0.000 | 0.037 | 0.000 | 0.069 | 0.000 |
| 7 | 0.045 | 0.000 | 0.017 | 0.000 | 0.096** | 0.000 |
| 8 | 0.004 | 0.000 | 0.007 | 0.000 | 0.117** | 0.000 |
| 9 | 0.052** | 0.000 | 0.007 | 0.000 | 0.010 | 0.000 |
| 10 | 0.054** | 0.000 | 0.036 | 0.000 | 0.017 | 0.000 |
| 11 | 0.021 | 0.000 | 0.445** | 0.000 | 0.121 | 0.000 |
| 12 | 0.034** | 0.000 | 0.270** | 0.000 | 0.047 | 0.000 |
| 13 | 0.017* | 0.010 | 0.073** | 0.000 | 0.009 | 0.000 |
| 14 | 0.003 | 0.004 | 0.006 | 0.000 | 0.043 | 0.000 |
| 15 | 0.001 | 0.000 | 0.017 | 0.000 | 0.010 | 0.000 |
| 16 | 0.007 | 0.000 | 0.021* | 0.000 | 0.007 | 0.000 |
| 17 | 0.118** | 0.000 | 0.001 | 0.000 | 0.021 | 0.000 |
| 18 | 0.028* | 0.000 | 0.009 | 0.000 | 0.150** | 0.000 |
| 19 | 0.126** | 0.000 | 0.000 | 0.000 | 0.048 | 0.000 |
| 20 | 0.066** | 0.000 | 0.214** | 0.000 | 0.149* | 0.000 |
| 21 | 0.040** | 0.000 | 0.013 | 0.000 | 0.003 | 0.000 |
| 22 | 0.001 | 0.000 | 0.040** | 0.000 | 0.002 | 0.000 |
| 23 | 0.002 | 0.000 | 0.079** | 0.000 | 0.221* | 0.000 |
| 24 | 0.003 | 0.000 | 0.188** | 0.000 | 0.289** | 0.011 |
| 25 | 0.021* | 0.000 | 0.003 | 0.000 | 0.024 | 0.000 |
| 26 | 0.037 | 0.000 | 0.044 | 0.000 | 0.201 | 0.000 |
| 27 | 0.005 | 0.000 | 0.011 | 0.000 | 0.091* | 0.000 |
| 28 | 0.021 | 0.000 | 0.036* | 0.000 | 0.101** | 0.000 |
| 29 | 0.469** | 0.000 | 0.131 | 0.000 | 0.030 | 0.000 |
| 30 | 0.019* | 0.000 | 0.032 | 0.000 | 0.000 | 0.000 |
| 31 | 0.014 | 0.000 | 0.135** | 0.000 | 0.430** | 0.000 |
| 32 | 0.009 | 0.000 | 0.087** | 0.000 | 0.005 | 0.000 |
Table 3: The proportional variance components of genotypic effects (VG/VP) and genotype-by-environment interaction effects (VGE/VP) for three wheat traits subject to Model 3.
Conclusion
In this study, two different models were employed to identify desirable DNA markers with high performance stability in winter wheat cultivars. The first model was a single marker model under each environment and the second one with GE effects included. The results obtained by both methods were comparable. Markers with major contributions to these three traits were identified by both models; however, GE interactions for these markers under these diverse environments were very small (Figure 1 and Table 3). DNA marker Rht1 on the short arm of chromosome 4B [54] was significantly detected for all three traits. This DNA marker was reported to be associated with plant anatomy and morphology [55,56]. Rht1 was also reported to be associated with Septoria tritici blotch [57]. It seems that Rht1 shows pleiotropic effects [58-61]. We also detected that the DNA marker VRN-D3-F6R8NED was highly associated with heading date [51]. The marker VRN-D3-F6R8NED is a vernalization gene, which controls heading date [51]. With the application of linear mixed model-based association mapping, in addition to DNA marker Rht1, DNA markers PPO18NED is associated with grain yield [62], Lr34JagTM is a wheat leaf rust resistance gene and was reported to be associated with plant growth and grain yield in Barley [63,64]. We found the similar result that this marker was significantly associated with plant height. Waxy-A1-AFC-AR2FAM, a co-dominant marker located on chromosome 8AS, was significantly associated with for heading date. Our results showed that the expression levels of the DNA marker Rht1 varied among different environments by single marker model and were confirmed by Model 2 as well.
References
- Brenchley R, Spannagl M, Pfeifer M, Barker GL, D'Amore R, et al. (2012) Analysis of the bread wheat genome using whole-genome shotgun sequencing. Nature 491: 705-710.
- UN Food and Agriculture Organization (FAO) (2013) Production of wheat by countries.
- Braun HJ, Atlin G, Payne T (2010) Multi-location testing as a tool to identify plant response to global climate change. Reynolds, CRP (Ed) Climate change and crop production, CABI, London, UK
- Donnelly P (2008) Progress and challenges in genome-wide association studies in humans. Nature 456: 728-731.
- Hayes B, Goddard M (2010) Genome-wide association and genomic selection in animal breeding. Genome 53: 876-883.
- Hayes BJ, Bowman PJ, Chamberlain AJ, Savin K, van Tassell CP, et al. (2009) A validated genome wide association study to breed cattle adapted to an environment altered by climate change. PloS ONE 4: e6676
- Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, et al. (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A 106: 9362-9367
- Pasam RK, Sharma R, Malosetti M, van Eeuwijk FA, Haseneyer G, et al (2012) Genome-wide association studies for agronomical traits in a worldwide spring barley collection. BMC Plant Biol 12: 16
- Wang M, Jiang N, Jia T, Leach L, Cockram J, et al (2012) Genome-wide association mapping of agronomic and morphologic traits in highly structured populations of barley cultivars. TAG Theoretical and applied genetics Theoretische und angewandte Genetik 124: 233-246
- Efron B, Hastie T, Johnstone I, Tibshirani R (2004) Least angle regression. Ann Stat 32: 407-451
- Hastie T, Tibshirani R, Friedman JH (2001) The elements of statistical learning: Data mining, inference and prediction. Springer, USA.
- Tibshirani R (1996) Regression shrinkage and selection via the Lasso. J Roy Stat Soc B Met 58: 267-288
- Breiman L (1984) Classification and regression trees. Wadsworth International Group, Belmont, Calif.
- Breiman L (2001) Random forests. Mach Learn 45: 5-32.
- Holland JH (1992) Adaptation in natural and artificial systems: An introductory analysis with applications to biology, control and artificial intelligence.
- Korte A, Vilhjalmsson BJ, Segura V, Platt A, et al. (2012) A mixed-model approach for genome-wide association studies of correlated traits in structured populations. Nat Genet 44: 1066-1071
- Segura V, Vilhjalmsson BJ, Platt A, Korte A, Seren U, et al. (2012) An efficient multi-locus mixed-model approach for genome-wide association studies in structured populations. Nat Genet 44: 825-830
- Stich B, Mohring J, Piepho HP, Heckenberger M, Buckler ES, et al (2008) Comparison of mixed-model approaches for association mapping. Genetics 178: 1745-1754
- Wang D, Zhu J, Li Z, Paterson A (1999) Mapping QTLs with epistatic effects and QTL × environment interactions by mixed linear model approaches. TAG Theoretical and applied genetics Theoretische und angewandte Genetik 99: 1255-1264
- Yu J, Pressoir G, Briggs WH, Vroh Bi I, Yamasaki M, et al. (2006) A unified mixed-model method for association mapping that accounts for multiple levels of relatedness. Nat Genet 38: 203-208
- Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, et al. (2010) Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42: 355-360
- Dodig D, Zoric M, Kobiljski B, Savic J, Kandic V, et al. (2012) Genetic and association mapping study of wheat agronomic traits under contrasting water regimes. Int J Mol Sci 13: 6167-6188.
- Le Couviour F, Faure S, Poupard B, Flodrops Y, Dubreuil P, et al. (2011) Analysis of genetic structure in a panel of elite wheat varieties and relevance for association mapping. Theor Appl Genet 123: 715-727
- Li FQ, Peng JH (2014) Genetic and association mapping study of english grain aphid resistance and tolerance in bread wheat germplasm. J Integr Agr 13: 40-53
- Mir RR, Kumar N, Jaiswal V, Girdharwal N, Prasad M, et al (2012) Genetic dissection of grain weight in bread wheat through quantitative trait locus interval and association mapping. Mol Breeding 29: 963-972
- Reif JC, Gowda M, Maurer HP, Longin CF, Korzun V, et al. (2011) Association mapping for quality traits in soft winter wheat. Theor Appl Genet 122: 961-970
- Wang M, Li Q, Xu L, Zhao J, Ma J, et al (2009) High-throughput genetic analysis and association mapping to identify novel genes for resistance to stripe rust in spring wheat germplasm. Phytopathology 99: S137-S137.
- von Korff M, Leon J, Pillen K (2010) Detection of epistatic interactions between exotic alleles introgressed from wild barley (H. vulgare ssp. spontaneum). TAG Theoretical and applied genetics Theoretische und angewandte Genetik 121: 1455-1464
- Xu S (2007) An empirical Bayes method for estimating epistatic effects of quantitative trait loci. Biometrics 63: 513-521
- Xu S, Jia Z (2007) Genomewide analysis of epistatic effects for quantitative traits in barley. Genetics 175: 1955-1963
- Zhou H, Steffenson B (2013) Genome-wide association mapping reveals genetic architecture of durable spot blotch resistance in US barley breeding germplasm. Mol Breeding 32: 139-154
- Agrama HA, Eizenga GC, Yan W (2007) Association mapping of yield and its components in rice cultivars. Mol Breeding 19: 341-356.
- Borba TCD, Brondani RPV, Breseghello F, Coelho ASG, Mendonca JA, et al. (2010) Association mapping for yield and grain quality traits in rice (Oryza sativa L.). Genet Mol Biol 33: 515-U142.
- Bryant R, Proctor A, Hawkridge M, Jackson A, Yeater K, et al. (2011) Genetic variation and association mapping of silica concentration in rice hulls using a germplasm collection. Genetica 139: 1383-1398.
- Famoso AN, Zhao K, Clark RT, Tung CW, Wright MH, et al. (2011) Genetic architecture of aluminum tolerance in rice (Oryza sativa) determined through genome-wide association analysis and QTL mapping.
- Li YH, Smulders MJM, Chang RZ, Qiu LJ (2011) Genetic diversity and association mapping in a collection of selected Chinese soybean accessions based on SSR marker analysis. Conserv Genet 12: 1145-1157
- Lu HY, Liu XF, Wei SP, Zhang YM (2011) Epistatic association mapping in homozygous crop cultivars. PLoS ONE 6: e17773
- Zhang WK, Wang YJ, Luo GZ, Zhang JS, He CY, et al. (2004) QTL mapping of ten agronomic traits on the soybean (Glycine max L. Merr.) genetic map and their association with EST markers. Theor Appl Genet 108: 1131-1139
- Shen X, Zhang T, Guo W, Zhu X, Zhang X (2006) Mapping fiber and yield QTLs with main, epistatic and QTL × environment interaction effects in recombinant inbred lines of upland cotton. Crop Sci 46: 61-66
- Zhao YL, Wang HM, Chen W, Li YH (2014) Genetic structure, linkage disequilibrium and association mapping of verticillium wilt resistance in elite cotton (Gossypium hirsutum L.) germplasm population.
- Arif MAR, Nagel M, Neumann K, Kobiljski B, Lohwasser U, Borner A (2012) Genetic studies of seed longevity in hexaploid wheat using segregation and association mapping approaches. Euphytica 186: 1-13.
- Breseghello F, Sorrells ME (2006) Association mapping of kernel size and milling quality in wheat (Triticum aestivum L.) cultivars. Genetics 172: 1165-1177.
- Zanke CD, Ling J, Plieske J, Kollers S, Ebmeyer E, et al (2014) Whole genome association mapping of plant height in winter wheat (Triticum aestivum L.).
- Peng J, Richards DE, Hartley NM, Murphy GP, Devos KMe (1999) 'Green revolution' genes encodes mutant gibberellin response modulators. Nature 400: 256-261
- Liu SY, Rudd JC, Bai GH, Haley SD, Ibrahim AMH, et al. (2014) Molecular markers linked to important genes in hard winter wheat. Crop Sci 54: 1304-1321
- Wu J (2014) Minque: An R package for linear mixed model analyses. R package version 10
- Wu J, Bondalapati K, Glover K, Berzonsky W, Jenkins JN, et al (2013) Genetic analysis without replications: Model evaluation and application in spring wheat. Euphytica 190: 447-458
- Zhu J (1989) Estimation of genetic variance components in the general mixed model. PhD thesis, North Carolina State University, Raleigh, USA.
- R Development Core Team (2010) R: A language and environment for statistical computing, R Foundation for Statistical Computing, Austria.
- Borrell AK, Incoll LD, Dalling MJ (1993) The influence of the Rht1 and Rht2 alleles on the deposition and use of stem reserves in wheat. Ann Bot Lond 71: 317-326.
- Chen YH, Carver BF, Wang SW, Cao SH, Yan LL (2010) Genetic regulation of developmental phases in winter wheat. Mol Breeding 26: 573-582.
- Jahan MA, Hossain MS, Khalekuzzaman M, Hassan MM (2007) Comparative genetic effect of dwarfing genes on yield and yield contributing traits in bread wheat. Progress in Agriculture 18: 49-55
- Storlie EW, Xie H, Talbert LE (1996) Tall off-types in semi dwarf spring wheat with height-reducing genes Rht1 and Rht2. Crop Sci 36: 1521-1522
- Somers DJ, Isaac P, Edwards K (2004) A high-density microsatellite consensus map for bread wheat (Triticum aestivum L.). Theor Appl Genet 109: 1105-1114
- Worland AJ, Korzun V, Roder MS, Ganal MW, Law CN (1998) Genetic analysis of the dwarfing gene Rht8 in wheat. Part II. The distribution and adaptive significance of allelic variants at the Rht8 locus of wheat as revealed by microsatellite screening. Theor Appl Genet 96: 1110-1120
- Worland AJ, Sayers EJ (1995) Rht1 (B dw), an alternative allelic variant for breeding semi-dwarf wheat varieties. Plant Breeding 114: 397-400.
- Baltazar BM, Scharen AL, Kronstad WE (1990) Association between dwarfing genes ‘Rht1’ and ‘Rht2’ and resistance to Septoria tritici Blotch in winter wheat (Triticum aestivum L. em Thell). Theor Appl Genet 79: 422-426.
- Borner A, Lehmann CO, Mettin J, Plaschke J, Schlegel G, et al. (1991) GA-insensitivity of ‘Aibian 1a’: pleiotropic effects of isogenic Rht-lines. Ann Wheat Newsletter 37: 59-60.
- Borrell AK, Incoll LD, Dalling MJ (1991) The Influence of the Rht1 and Rht2 Alleles on the growth of wheat stems and ears. Ann Bot Lond 67: 103-110.
- Kertesz Z, Flintham JE, Gale MD (1991) Effects of Rht dwarfing genes on wheat grain yield and its components under Eastern European conditions. Cereal Res Commun 19: 297-304
- Nizam UM, Marshall DR (1989) Effects of dwarfing genes on yield and yield components under irrigated and rain fed conditions in wheat (Triticum aestivum L.). Euphytica 42: 127-134
- Sun DJ, He ZH, Xia XC, Zhang LP, Morris CF, et al (2005) A novel STS marker for polyphenol oxidase activity in bread wheat. Mol Breeding 16: 209-218
- Risk JM, Selter LL, Chauhan H, Krattinger SG, Kumlehn J, et al (2013) The wheat Lr34 gene provides resistance against multiple fungal pathogens in barley. Plant Biotechnol J 11: 847-854
- Risk JM, Selter LL, Krattinger SG, Viccars LA, Richardson TM (2012) Functional variability of the Lr34 durable resistance gene in transgenic wheat. Plant Biotechnol J 10: 477-487.