Research Article, J Appl Bioinforma Comput Biol Vol: 10 Issue: 5
Neglected Disdb: A Broad Internet Framework for Gathering And Analysing Data From Neglected Diseases
Venu Paritala*, Rajashekhar Reddy S and Sukesh Kalva
Department of Biotechnology, Vignan’s Foundation for Science, Technology & Research (Deemed to be University), Vadlamudi, Guntur, Andhra Pradesh, India
*Corresponding Author: Venu Paritala
Department of Biotechnology, Vignan’s Foundation for Science, Technology & Research (Deemed to be University), Vadlamudi, Guntur, Andhra Pradesh, India
E-mail: vvenuparitala@gmail.com
Received: April 19, 2021 Accepted: May 19, 2021 Published: May 26, 2021
Citation: Paritala V, Reddy RS, Kalva S (2021) Neglected Disdb: A Broad Internet Framework for Gathering And Analysing Data From Neglected Diseases. J Appl Bioinforma Comput Biol 10:5.
Abstract
Biological databases, which offer access to a wide range of biologically related data, play a vital role in the life sciences. This paper suggests a community-defined, uniform, generic definition of the biological database’s core quality. This paper is mostly intended for computer scientists with only a basic knowledge of biology. Scientific literature is being replaced by libraries, Understand the details crowded in statistics and text as a way of disseminating this information to the public neglected disease database (https:// venuparitala.shinyapps.io/Neglated-diseasedb/). The NEGLECTED DISDB is a biological archive that includes disease knowledge for about 20 neglected diseases, as well as genes, targets, and gene network analysis. This database is open to the public, free, and open-source. it was developed with r programming language in r studio by using different packages.
Keywords: Network; genes; targets; database and disease
Introduction
Neglected diseases are a category of diseases that flourish in areas with inadequate access to proper hygiene, clean water, and healthcare, as well as areas where humans live in close proximity to livestock and disease vectors, such as remote and rural areas, informal settlements (or) hot spots. Neglected diseases mainly impact people living in poverty and the most marginalised sections of society in Africa, Asia, and the United States. Around the world, untreated tropical diseases impact over a billion people. Every year, 270 million children under the age of three and over 600 million school-aged children are impacted by malnutrition and disfigurement and social decrimination. Both the ability to recognise the immune processes during parasite configuration in the host and the ability to manage and remove ntd are necessary for control and elimination. As a result, a database is created for the following ntds, which are described on the World Health Organization's website, as well as CDs websites created using the r programming language in r studio using various packages.
1.Buruli Ulcer 2. Chagas Disease 3.Cysticercosis 4.Dengue Fever 5.Dracunculiasis (Guinea Worm Disease) 6.Echinococcosis 7.Fascioliasis 8.Human African Trypanosomiasis (African Sleeping Sickness) 9.Leishmaniasis 10.Leprosy (Hansen’s Disease) 11.Lymphatic Filariasis 12.Mycetoma 13.Onchocerciasis 14.Rabies 15.Schistosomiasis 16.Soil-transmitted Helminths (STH) (Ascaris, Hookworm, and Whipworm) 17.Trachoma 18.Yaws 19.Taeniasis and neurocysticercosis 20. Foodborne trematodes.
This database provides extensive details about any neglected illness, including species, genes, network plots, effects, and disease prevention, as well as information from PubMed (https://pubmed. ncbi.nlm.nih.gov/). This data will be used by bioinformaticians and biomedical researchers to design drugs and Insilco vaccines, the protein targets responsible for the neglected disease are retrieved from the ChemBL database(https://www.ebi.ac.uk/chembl/), for further studies in drug discovery [1].
This database provides extensive details about any neglected illness, including species, genes, network plots, effects,
Methods
Implementation
NEGLECTED DISDB web application is developed with R (Reproducible Research with R and R Studio, Second Edition, 2018). The server is hosted in a cloud platform. For the development of the analytical pipeline, various R packages were used [2]. The packages are shiny, shiny dashboard, shiny themes, dashboard themes, slickR, DT, gg plot2, multicolor, gg network, shiny css loaders, shiny custom loader, html tools, html widgets. It contains both the user interface and server.
Shiny: Shiny is a R kit that allows you to build immersive web apps directly from R. You can host standalone applications on a website, insert them in R Markdown documents, or build dashboards using R Markdown. You can also add CSS themes, html widgets, and JavaScript actions to your Shiny applications.
Shiny dashboard: shiny dashboard is an R package whose job is to make it easier, as the name suggests, to build dashboards with Shiny
Shiny themes: used top beatification of web page.
Dashboard themes: Customise the Appearance of ‘shiny dashboard’ Applications using Themes. Allows manual creation of themes and logos to be used in applications created using the ‘shiny dashboard’ package. Removes the need to change the underlying css code by wrapping it into a set of convenient R functions.
Slickr: Create Interactive Carousels with the ‘JavaScript’ ‘Slick’ Library. Create and customize interactive carousels using the ‘Slick’ ‘JavaScript’ library and the ‘html widgets’ package. The carousels can contain plots produced in R, images, ‘iframes’, videos and other ‘html widgets’.
DT: the r package provides an r interface to the javascript library data tables. r data objects (matrices or data frames) tin be displayed as tables on HTML pages, and data tables provide filtering, pagination, sorting, and many other features in the tables.
ggplot2: ggplot2 is a R package dedicated to data visualization. It can greatly improve the quality and aesthetics of your graphics, and will make you much more efficient in creating them.
Multicolor: Add Multiple Colors to your Console & R Markdown Output. Add multiple colors to text that is printed to the console.
Ggnetwork: The ggnetwork package provides a way to build network plots with ggplot2. Gg network further requires the network.
Shiny css loaders: shiny css loaders Add Loading Animations to a ‘shiny’ Output While It’s Recalculating. When a ‘Shiny’ output (such as a plot, table, map, etc.) is recalculating, it remains visible but gets greyed out.
Shiny css loaders: Instead, you can use’ shiny css loaders’ to apply a loading animation (“spinner”) to outputs. A spinner can appear when the output is recalculating if you wrap a ‘Shiny’ output in ‘with Spinner()’.
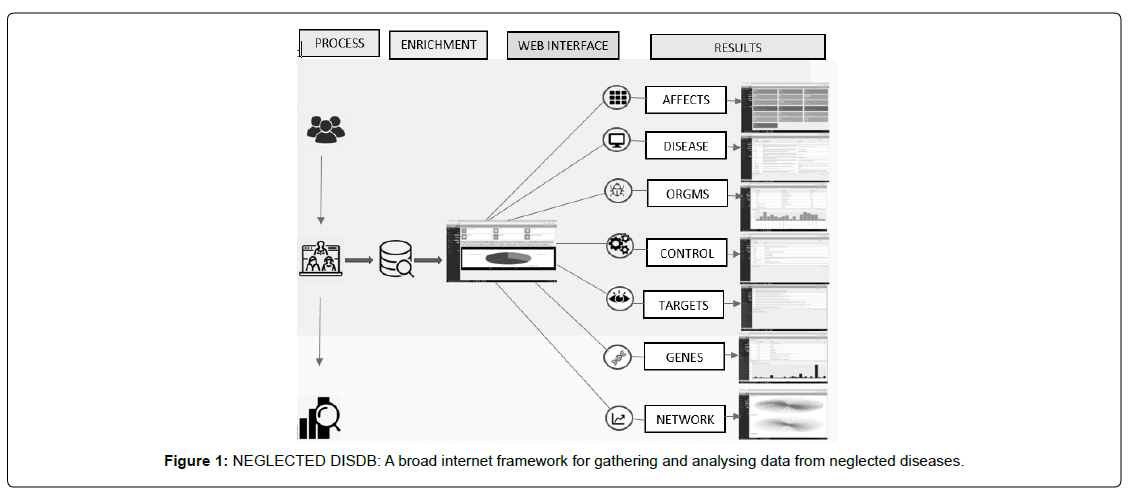
Html tools: htmltools [3] is a R package designed to: Generate HTML tags from R. Handle web dependencies [figure 1].
Html widgets: The package provides a framework for easily creating R bindings to JavaScript libraries. Widgets created using the framework can be: Used at the R console for data analysis just like conventional R plots (via R Studio Viewer).
Result and Discussion
This database contains the complete information of 20 neglected diseases including the disease tab panel (disease information, disease name), effects tab panel (shows the data how many people are affected), organism tab panel (disease name, disease-causing organism, organism type), control panel (disease name and control disease information), target panel (chembl id; “name”; “UniProt accessions”; “type”; “organism”; “compounds”; “activities”; “tax id”; “species group), each disease having gene name, gene symbol, and frequency present in the gene tab panel, finally pathway and network analysis graphical representation are plotted in the network tab panel.
Home: It is a dashboard page divided into two partitions, one hand represents the database, disease images. On the other hand, there is an overall collection of database representation.
Affect: This tab panel contains information about statistics of diseased population by each neglected disease. On the left hand of the application click on the disease tab panel to explore disease information at another hand.
Disease: This tab panel contains the disease information, and symptoms of each neglected disease out of all neglected diseases, it presents the top filters which easily. On the left hand of the dashboard page click on the disease tab panel to explore disease information on another hand.
Organism: This component represents organisms responsible for disease and it shows the type of organisms like bacteria, virus, fungi, protozoa, parasite, tapeworm, etc., it plots a graph between organism type and disease which represents which disease is affecting more for eg: virus is more vital than others. User can search for a specific disease in the search box given
Control: tab panel consists of information about which drugs/ prescriptions to be taken for all neglected diseases. On the left hand of the dashboard page click on the disease tab panel to explore disease information on another hand. It presents the top filters which easily to search specific disease name.
Targets: Targets tab panel represents all diseases when you select a disease its display’s all possible targets with accssion id compounds [4, 6].
Genes: This genes tab panel represents all diseases when you select a disease it displays all possible gene names with gene symbol and frequency. On the left hand of the dashboard page click on the disease tab panel to explore about/gene information on another hand. it presents the top filters which easily search specific gene name. It plots a graph between gene symbol and frequency which represents which disease is affecting more for eg: T code 30 times is more vital than others.
Network: This tab panel contains the network connection between the genes data. It displays the pathway analysis to find [7,8].
Conclusion
This database presents overall and complete information regarding neglected diseases, it is free, open-source, and user-friendly, it portable/compatible on any operating system.
Funding
The author(s) received no financial support for the research.
Acknowledgment
Author thanks DST FIST facility provided by the Vignan’s institute for science, technology and research for execution of work.
Conflict of interest
None
Copyright
This work is registered by copyright authority of India. (ROC :SW-13927/2020).
References
- Neglected tropical disease [Internet] available from 1. https://www.who.int/teams/control-of-neglected-tropical-diseases
- GBD (2016) “Disease and Injury Incidence and Prevalence Collaborators (2017) Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016”. Lancet 390: 1211–59.
- Herricks JR, Hotez PJ, Wanga V, Coffeng LE, Haagsma JA, et al. (2017) “The Global Burden of Disease Study 2013: What does it mean for the NTDs?” PLoS Negl Trop Dis 11: 005424.
- Hotez PJ, Pecoul B, Rijal S, Boehme C, Aksoy S, et al. (2016) “Eliminating the Neglected Tropical Diseases: Translational Science and New Technologies”. PLoS Negl Trop Dis 10(3):3895.
- Brook CE, Beauclair R, Ngwenya O, Worden L, Ndeffo-Mbah M, et al. (2015) “Spatial heterogeneity in projected leprosy trends in India.” Parasit Vectors 8:542.
- Albonico M, Montresor A, Crompton DW, Savioli L (2006) “Intervention for the control of soil-transmitted helminthiasis in the community Adv.” Parasitol 61: 311-348.
- Amazigo UV, Obono M, Dadzie KY, Remme J, Jiya J et al. (2002) “Seketeli Monitoring community-directed treatment programmes for sustainability: lessons from the African Programme for Onchocerciasis Control (APOC) Ann.” Trop. Med. Parasitol 96: 75-92.
- Simon J B, Kundai Z, Nebiyu N, Siobhan C, Mona H. (2004) “Neglected tropical disease control in a world with COVID-19: an opportunity and a necessity for innovation.