Briefreport, Appl Bioinforma Comput Biol Vol: 12 Issue: 3
Omics Finder: An Android Application to Assist Pyhton and Omics Queries
A Jawaria, Syed Ali Raza Shah*, Mehrosh Khalid, Attiya Kanwal and Tahira Noor
Department of Bioinformatics, International Islamic University, Islamabad, Pakistan
*Corresponding Author: Syed Ali Raza Shah
Department of Bioinformatics, International Islamic University, Islamabad, Pakistan
E-mail: Zebishah15@yahoo.com
Received date: 29 May, 2023, Manuscript No. JABCB-23-100400;
Editor assigned date: 31 May, 2023, PreQC No. JABCB-23-100400 (PQ);
Reviewed date: 21 June, 2023, QC No. JABCB-23-100400;
Revised date: 28 June, 2023, Manuscript No. JABCB-23-100400 (R);
Published date: 05 July, 2023, DOI: 10.4172/2327-4360.100071
Citation: Jawaria A, Shah SAR, Khalid M, Kanwal A, Noor T (2023) Omics Finder: An Android Application to Assist Pyhton and Omics Queries. J Appl Bioinforma Comput Biol 12:3.
Abstract
Bioinformatics is a rapidly flourishing field with new innovations and technologies. In recent years, the development of novel and powerful bioinformatics applications dedicated to biological data acquisition empowered both the basic and applied life sciences research. Omics Finder is an offline educational android application that provides learning material and assistance in research related queries to the researchers, academicians and scientists in the field of life sciences in a fast and nice user-interface. The main characteristic of Omics Finder is the integration of domain specific information and mainly focuses on three research domains such as genomics, proteomics and python. Omics Finder can be used for book consultation, video lectures, notes, slides and FAQs to gain in depth knowledge about specific domains. This application comprise of domain specific queries and top cited books, lecture videos, notes, and slides that were taken from different platforms like Bio Star (https://www.biostars.org/), Stack Over Flow (https://stackoverflow.com/), Quora (https://www.quora.com/), Research Gate (https://www.researchgate.net/), YouTube (https://www.youtube.com/) and Google. In total, 300 unanswered queries were extracted from those platforms and 82 answered queries were added to Omics Finder. The user will interact with the application through mobile device and will avail the mediated and self-access services via getting access to prescribed services. It was implemented using Java programming language and it is not freely available for commercial use. Omics Finder is developed to help and assist early career researchers and professionals in their research studies by Providing: (i) Free platform (ii) Answer to the searched query (iii) Referenced answers (iv) Assistance material in several formats (Books, Video lectures, Notes, Slides and FAQs).
Keywords: Bioinformatics, Omics, Python, Android application
Introduction
Bioinformatics is the digitalization, acquisition, storage and analysis of biological data via computer based tools and to make the data more efficient to use. It has filled the gap between biology and information technology by bringing biological themes into computers [1,2]. The developed tools and applications are not just helping the scientists, researchers, biologists but also the students who have recently stepped in the field of bioinformatics. For the analysis of the research data, bioinformatics tools, applications or softwares are the need of hour which can provide fast, quick and authentic information and solutions [3].
Increased accessibility of smart phone has lead to the development and use of different freely available applications (health) behavior change, reading current health information, entertainment and socializing purposes, as well as making online purchases [4]. Mobile technology and educational applications supports the gaining of knowledge and skills in clinical settings. Moreover, researchers from different fields have reported various benefits of mobile technology and applications. For instance, quick access to educational materials, information about gene datasets, proteomics and python [5].
Mobile learning (M-learning) applications has the capability to expand education and it is the most convenient and quick ways to obtain academic materials [6]. Mobile applications are made with a specific intention, which are stored in mobile application store such as gaming applications with a goal of learning education, knowledge; promote problem-solving and cooperative skills [7]. Apart from gaming applications, many educational programs and technologies are available which can help students in learning process [8]. List of M-Learning Applications used for learning are presented in Table 1.
Table 1: List of Mobile learning applications used for learning.
The pandemic of COVID-19 has caused disruption in normal daily life which has led to the closure of schools, colleges and universities. Education system is highly disturbed due to facing difficulty in making policies for education [13]. Online Learning management software and open-source digital learning applications are adopted to ensure the continuity in institutes and universities for learning and teaching [14]. Online transformation affected the structure of learning, schooling, and teaching and assessment methodologies which caused many problems for the Students, teachers and parents [15].
Methodology
Planning
Inclusion: The inclusion criteria for the chosen queries in application includes (i) Queries in English language (ii) Domain specified queries (iii) Most top cited books, lecture videos, notes, and slides were selected.
Exclusion: The exclusion criteria for the chosen queries in application include (i) Queries in non-English language (ii) Nondomain specified queries (iii) Irrelevant domain and closed queries were excluded.
Information source: Different platforms like BioStar (https://www.biostars.org/) [16], StackOverFlow (https://stackoverflow.com/) [17], Quora (https://www.quora.com/) [18], Research Gate (https://www.researchgate.net/) [19], YouTube (https://www.youtube.com/) [20] and Google were visited to gather domain based queries and answers.
Searching procedure: Manual search approach was performed by visiting above mentioned (section 1.3) selected platforms for the collection of queries, referenced answers, books, video lectures, notes and slides.
Data extraction: Based on the inclusion criteria, the query data and other associated assistance material has been extracted by manual searching and selection process. More than 100 queries were extracted for each domain from selected platforms mentioned in (section 1.3). The domain specific relevant queries were selected. A total of 82 unanswered queries (25 for genomics, 31 for python, and 26 for proteomics), Top rated books, slides, notes and lecture videos from different platforms were selected and added in database.
Development: Omics-Finder was implemented using Java programming language for the System development and the functionalities. Queries, referenced answers, reference for answer source, link for book, slide, notes and lecture video references were stored in realtime firebase database as described by Moroney [21]. Android studio, Software Development Kit (SDK) tool was used for the implementation of functionality into the system and application development [22].
Working: The basic workflow of application is depicted in Figure 1.
Result Outputs
The home page of Omics Finder is followed by the authentication process that will take the user to main page of Omics Finder. The Main page will show two search options (i) Query search (ii) Domain search. Query search option will allows the user to search for query by inserting the keyword match search and the list of relevant queries will be displayed to the user. Each query will lead the user to referenced answer, reference and reference link. However, domain search option provides domain wise assistance to the user by displaying list of books, videos, notes, slides and domain relevant FAQs. The systems outputs are depicted in Figure 2-5.
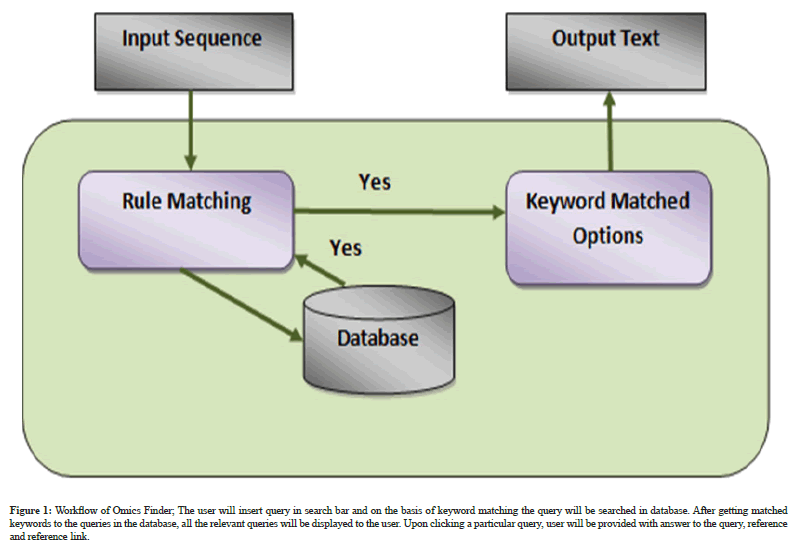
Figure 1: Workflow of Omics Finder; The user will insert query in search bar and on the basis of keyword matching the query will be searched in database. After getting matched keywords to the queries in the database, all the relevant queries will be displayed to the user. Upon clicking a particular query, user will be provided with answer to the query, reference and reference link.

Figure 2: Home Page of Omics Finder; To get started with the application, the home page of OmicsFinder is followed by the authentication process. After completing the registration, user will be directed to the mainpage of OmicsFinder where two search options will be displayed.
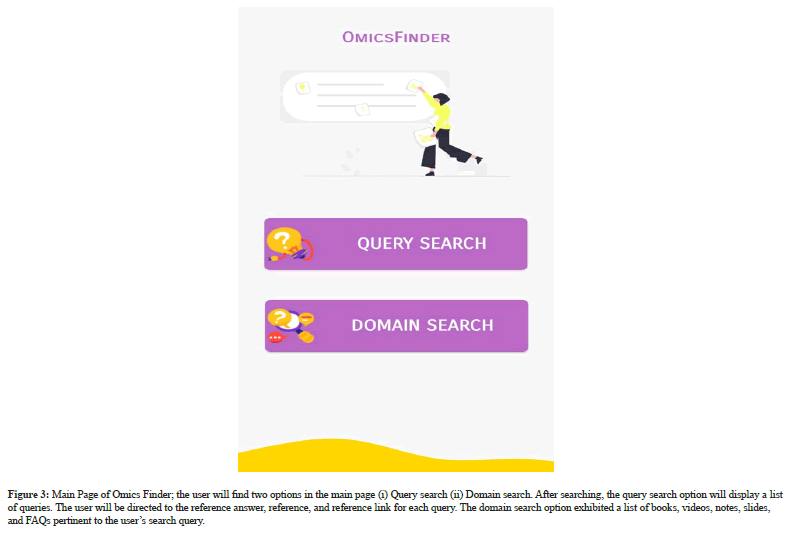
Figure 3: Main Page of Omics Finder; the user will find two options in the main page (i) Query search (ii) Domain search. After searching, the query search option will display a list of queries. The user will be directed to the reference answer, reference, and reference link for each query. The domain search option exhibited a list of books, videos, notes, slides, and FAQs pertinent to the user’s search query.
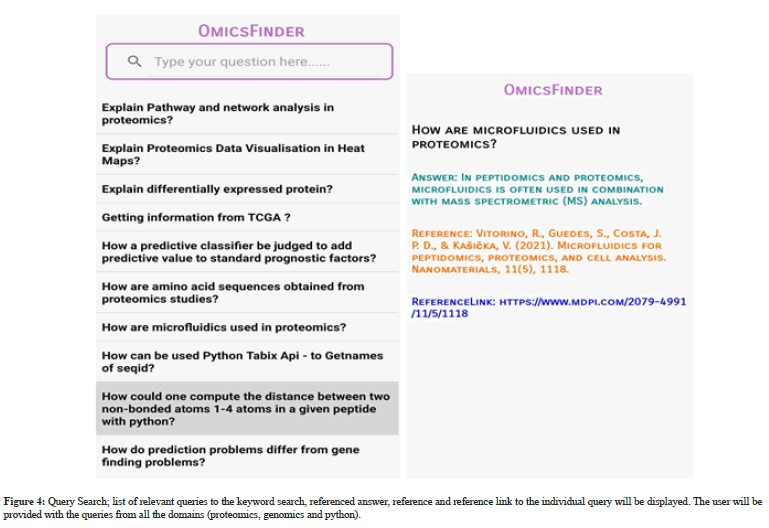
Figure 4: Query Search; list of relevant queries to the keyword search, referenced answer, reference and reference link to the individual query will be displayed. The user will be provided with the queries from all the domains (proteomics, genomics and python).
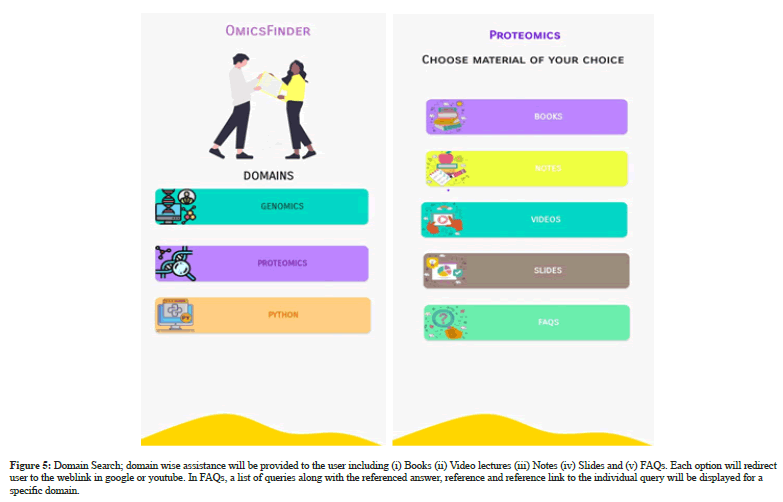
Figure 5: Domain Search; domain wise assistance will be provided to the user including (i) Books (ii) Video lectures (iii) Notes (iv) Slides and (v) FAQs. Each option will redirect user to the weblink in google or youtube. In FAQs, a list of queries along with the referenced answer, reference and reference link to the individual query will be displayed for a specific domain.
Discussion
The area of bioinformatics is quickly growing with new advancements and technologies. Researchers, academicians and scientists need assistance in their research and studies in parallel to cope up with scientific information and innovations. Technology makes the learning process more interactive and interesting. Android and smartphone technology can be used as learning medium to provide efficient and effective learning material applications [23]. Omics Finder is a mobile application for python and omics queries. It is developed as a solution to bridge the gap for proteomics, genomics and python knowledge through user queries, books, lecture videos, lecture notes and slides and provide assistance in the field of life sciences and computer science. Different applications are available such as SoloLearn [24,25], MAC Ben Abim [26] and BengaWolex [27]. SoloLearn and Milo are programming based android applications and provides visual programming environment to learn coding and several computer programming languages for the data science education, computer science education, applied mathematics and algebra [28], while Benga Wolex and MACBenAbim are multi-agentbased cross-platform mobile application for Omics, Computational Biology and Bioinformatics [29]. In biological sciences, omics refers to the study of genomics, proteomics and transcriptomics. Genomics is the study of genome or genes and proteomics refers to the study of all the proteins in cell, tissue or organism [30]. Python is very flexible, user-friendly, high-level, dynamic, object-oriented, general purpose programming language that is used in every field of science especially for the analysis of biological and computational problems [31]. However, Omics Finder helps in the domains of both Omics (proteomics, genomics) and python programming. The user will get allowance to those services by registering to the applications but these applications along with Omics Finder are yet unavailable for open and commercial use.
Conclusion
Mobile devices have been used effectively to extend the learning environment. Omics Finder is developed to help and assist early career researchers and professionals in their research studies by Providing free platform, response to the searched query, and referenced answers along with Assistance material in different formats like books, video lectures, notes, slides and FAQs. In near future, more research domains and new modifications will be added and will be given access for free to all the scientific fraternity.
Limitations
The limitation of Omics Finder applications is that only three domains of research were targeted such as Python, Genomics and Proteomics. Each domain has a huge set of queries but limited set of queries, authentic referenced answers and assistance material was added to the application. These limitations can be overcome by performing new modifications and adding more research domains to the application in near future.
References
- Chen H, Yu T, Chen JY (2013) Semantic web meets integrative biology: A survey. In Briefings in Bioinformatics. Brief Bioinform 14:109-125.
[Crossref] [Google Scholar] [Pubmed]
- Inza I, Calvo B, Armañanzas R, Bengoetxea E, Larrañaga P et al. (2010) Machine learning: an indispensable tool in bioinformatics. Methods mol biol 593:25-48.
[Crossref] [Google Scholar] [Pubmed]
- Dash S, Shakyawar SK, Sharma M, Kaushik S (2019) Big data in healthcare: management, analysis and future prospects. J Big Dat 6:54.
- Fiona H M, Christina C, Anemarie W, Jane S, Huge S et al. (2018) Evaluating mobile phone applications for health behaviour change: A systematic review. J Telemed Telecare 24: 22-30.
[Crossref] [Google Scholar] [Pubmed]
- O’Connor S, Andrews T (2018) Smartphones and mobile applications (apps) in clinical nursing education: A student perspective. Nurse Educ Today 69:172-178.
[Crossref] [Google Scholar] [Pubmed]
- Sarrab M (2012) Mobile Learning (M-Learning) and Educational Environments. Int J Distrib Syst Parallel 3: 31-38.
- Connolly T M, Stansfield M, & Hainey T (2011) An alternate reality game for language learning: ARGuing for multilingual motivation. Comput Educ 57: 1389-1415.
- Drigas AS, Angelidakis P (2017) Mobile applications within education: An overview of application paradigms in specific categories. Int J Interact Mob Technol 11:17- 29.
- Van Arnhem, Pieta J (2013) Unpacking Evernote: Apps for Note-Taking and a Repository for Note-Keeping. Charleston Adv 15:55-57.
- Shi Y, Tsai C (2022) Fostering vocabulary learning: mind mapping app enhances performances of EFL learners. Comput Assist Lang Learn.
- Gudmundsson V, Lindvall M, Aceto L, Bergthorsson J, Ganesan, D (2016) Model-based testing of mobile systems-An empirical study on QuizUp Android app 208:16-30 Electron Proc Theor Comput Sci.
- Liu C, Correia AP (2021) A case study of learners’ engagement in mobile learning applications. Online Learn J 25:25-48.
- Dutta S, Smita M K (2020) The Impact of COVID-19 Pandemic on Tertiary Education in Bangladesh: Students’ Perspectives. Open J Soc Sci 8:53-68.
- Ferri F, Grifoni P, Guzzo T (2020) Online learning and emergency remote teaching: Opportunities and challenges in emergency situations. Societies.
- Bhasin B, Gupta G, Malhotra S (2021) Impact of COVID-19 pandemic on education system. EPRA ECEM. 8: 6-8.
- Parnell LD, Lindenbaum P, Shameer K, Dall’Olio GM, Swan DC, et al. (2011) BioStar: An online question & answer resource for the bioinformatics community. PLoS Comput Biol 7:1-5.
[Crossref] [Google Scholar] [Pubmed]
- Allamanis M, Sutton C (2013) Why, when, and what: Analyzing stack overflow questions by topic, type, and code. IEEE 53-56.
- Noviyanti S D (2023) Student’s Perspective of Using Quora: An Authentic Learning Experience in Digital Platform. J Eng Lang Teach Appl Linguist 4:45- 51.
- Ovadia S (2014) Research Gate and Academia.edu: Academic Social Networks Behav Soc Sci Librar 33:165-169.
- Snelson C (2011) YouTube across the Disciplines: A Review of the Literature. J Online Learn Teach 7:159-169.
- Moroney L (2017) The Firebase Realtime Database: In The Definitive Guide to Firebase. 51-71
- Esmaeel HR (2015). Apply Android Studio (SDK) Tools. Int j adv res comput sci softw eng 5:88-93.
- Nofitasari A, Lisdiana L, Marianti A (2021) Development of My Biology App Learning Media Based On Android Materials of Food Digestion Systems as Student Learning Source at MA. J Innov Sci Educ 10(1): 70-78.
- Quinn A (2018) Learning to Program with SoloLearn. The Mathematics Teacher 112:226-230.
- Rao A, Bihani A, Nair M (2018) Milo: A visual programming environment for data science education. IEEE 18: 211-215.
- Oluwagbemi O, Adewumi A, Esuruoso A (2012) MACBenAbim: A Multi-platform Mobile Application for searching keyterms in Computational Biology and Bioinformatics. J Bioinform 8(16): 790–791.
[Crossref] [Google Scholar] [Pubmed]
- Oluwagbemi O, Adewumi A, Esuruoso A (2013) BengaWolex: A Novel Cross-platform mobile application for storing and accessing fundamental knowledge in transcriptomics, proteomics, metagenomics and metabolomics. J Bioinform 8:790-791.
- Maloney J, Resnick M, Rusk N, Silverman B, Eastmond E (2010) The scratch programming language and environment. ACM Trans Comput Educ 16:1-15.
- Deja M, Rak, D, Bell B (2021) Digital transformation readiness: perspectives on academia and library outcomes in information literacy. J Acad Librariansh 47(5): 1-15.
- Vailati Riboni, Palombo V, Loor JJ (2017). What are omics sciences? In Periparturient Diseases of Dairy Cows: A Systems Biology Approach 1-7.
- Sharma A, Khan F, Sharma D, Gupta S, Student FY (2020) Python: The Programming Language of Future Int J Innov Res Technol 6:115- 118.

