Case Report, J Appl Bioinforma Comput Biol Vol: 7 Issue: 4
Specific Characteristics of the Esophageal Carcinosarcoma at Multi-Omics Levels: A Rare Case Report
Qian Yang1, Jian-Zhong He1, Shao-Hong Wang2, Shang Li3, Yang Chen1, Zhi-Yong Wu2, Shuai-Xia Yu1, Xiu-E Xu1, Chun-Quan Li4, En-Min Li1* and Li-Yan Xu1
1The Key Laboratory of Molecular Biology for High Cancer Incidence Coastal Chaoshan Area, Shantou University Medical College, Shantou 515041, China
2Shantou Central Hospital, Affiliated Shantou Hospital of Sun Yat-sen University, Shantou 515041, China
3The College of Bioinformatics Science and Technology, Harbin Medical University, Harbin 150081, China
4Department of Medical Informatics, Daqing Campus, Harbin Medical University, Daqing, 163319, China
*Corresponding Author: En-Min Li
The Key Laboratory of Molecular Biology for High Cancer Incidence Coastal Chaoshan Area, Shantou University Medical College, No. 22, Xinling Road, Shantou, Guangdong 515041, China
Tel: +86-754-8890-0460
Fax: +86-754-8890-0847
E-mail: nmli@stu.edu.cn
Received: November 11, 2018 Accepted: December 14, 2018 Published: December 24, 2018
Citation: Yang Q, He J, Wang S, Li S, Chen Y, et al. (2018) Specific Characteristics of the Esophageal Carcinosarcoma at Multi-Omics Levels: A Rare Case Report. J Appl Bioinforma Comput Biol 7:4. doi: 10.4172/2329-9533.1000159
Abstract
Esophageal carcinosarcoma is a rare cancer type, which accounts for approximately 3% in esophageal neoplasms with low 5-year overall survival. Carcinosarcoma is biphasic in nature which consists of both carcinomatous and sarcomatous elements. Of these sarcomatous components, fibrosarcoma is the most common type and others include leiomyosarcoma, rhabdomyosarcoma, chondrosarcoma, osteosarcoma, liposarcoma, chondrosarcoma and so on. Since the first description of this neoplasm by Chapman in 1877, esophageal carcinosarcoma cases have been reported, but its pathogenesis is still unclear. Here, we report a case with complete follow-up data and integrated high-throughput data, which could contribute to elucidating the pathophysiological mechanisms of diseases accurately and provide novel therapeutic strategies to the clinical research. To the best of our knowledge, this is the first reported case to characterize its characteristics at multi-omics levels.
Keywords: Esophageal carcinosarcoma; Immunohistochemistry; Multi-omics data; Bioinformatics; Precision medicine
Introduction
Esophagus cancer ranks sixth and eighth worldwide in cancer mortality and incidence, respectively [1]. The most common histological types are esophageal squamous cell carcinoma (ESCC) and esophageal adenocarcinoma (EAC). In addition, esophageal carcinosarcoma (EC, less than 3%) is a relatively rare malignant neoplasm in esophageal cancer with low 5-year overall survival [2]. EC often presents as an intraluminal polypoid lesion with dysphagia and characterized by an admixture of malignant epithelial components (carcinoma) and malignant mesenchymal components (sarcoma). Fibrosarcoma is one of a common type in EC, which is a malignant mesenchymal tumor derived from fibrous connective tissue with uncertain treatment and prognosis [2]. The treatment and prognosis of EC is still uncertain. Previous studies report that carcinosarcoma appears in patients with a history of chemotherapy or radiotherapy, but there is still no established therapeutic strategy for esophageal carcinosarcoma [3].
Recent studies have emphasized that integrating multi-omics data could improve the understanding of patient symptoms in multiple dimensions and explore novel therapeutic strategies for diseases in precision oncology [4]. Here, we report a rare case, carcinosarcoma and squamous cell carcinoma coexisting in the esophagus, and characterize its specific features based on the corresponding multiomics data, which could provide novel therapeutic strategies and promote the development of precision medicine.
Case Report
A 60-year-old man presented to the hospital for a further examination of his progressive dysphagia and weight loss in past one month. Laboratory values revealed that the levels of the serum tumor markers carcinoembryonic antigen (CEA) increased (6.46 ng/ml) and other laboratory data were within the normal limits. Esophagogastroduodenoscopy (EGD) shown that a polypoid neoplasm with ulceration which surface is uneven, located at 32- 36 cm from the incisors. No significant lymphadenopathy and metastatic lesions were detected by the image examination. The patient underwent left transthoracic esophagectomy in treatment of middle thoracic esophagus carcinoma without preoperative adjuvant therapy.
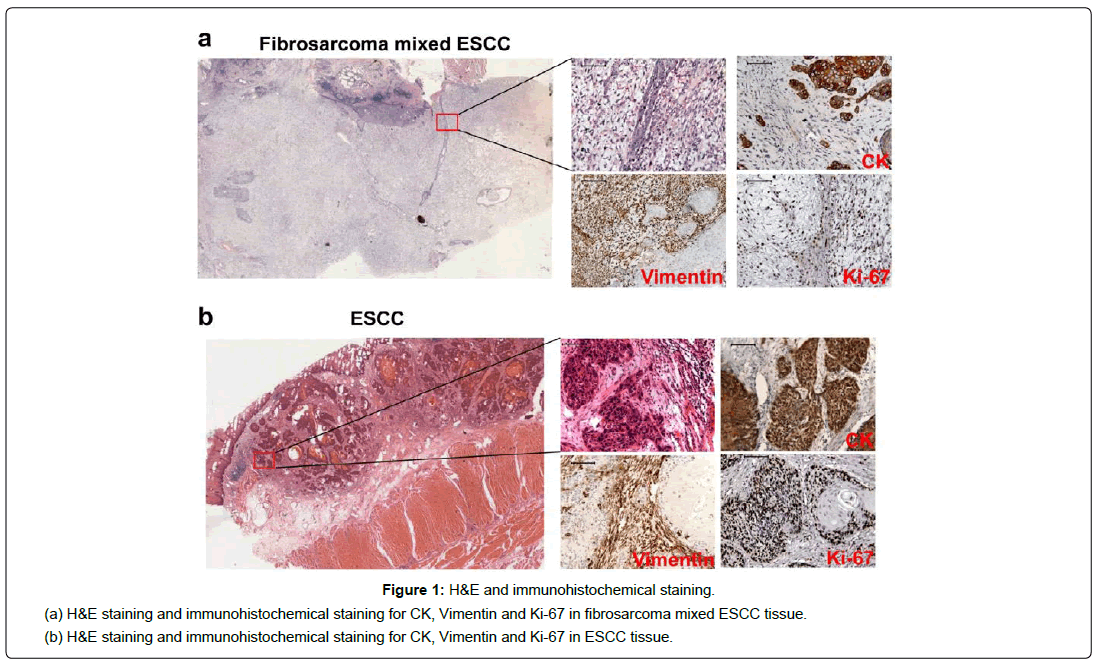
The histological finding revealed that the tumor consisted of two components: squamous cell carcinoma and sarcomatous element of spindles-shaped cells, and the transitional features between them were evident (Figure 1a). Immunohistochemical further analysis reveals that the squamous cell carcinoma cells were positive for CK (Figure 1a) and CK17, while the spindle cells were diffusely positive for vimentin (Figure 1a). The positive rate of Ki-67 for squamous cell carcinoma cells and spindle cells were 50% and 80%, respectively (Figure 1a). And the stains for Actin, Lysozyme and Myoglobin were negative. All lymph nodes (0/26) were negative for tumor. According to the WHO classification system the final pathologic diagnosis was carcinosarcoma: squamous cell carcinoma mixed fibrosarcoma, and the clinical diagnosis based on the seventh edition of the Union for International Cancer Control (UICC) was T3N0M0 Stage ⅡA. The patient had a smooth recovery and discharged after 10 days of surgery.
During subsequent follow-up, patient developed hoarseness and dysphagia four months after surgery. Typical CT examination appearance was wall thickening at the anastomotic site and the upper mediastinal. The serum tumor markers CEA increased to 7.85 ng/ml. A radiotherapy was given to the patient with a dose of 50 Gy (Partially increased to 64-66 Gy), and concurrent chemotherapy with Tegafur/Uracil. Ultimately, he was died from tumor local recurrence, extensive metastasis and pulmonary infection 9 months after operation. The study was approved by the ethical committee of the Central Hospital of Shantou City and the ethical committee of the Medical College of Shantou University, and a written informed consent was obtained from the patient to use resected samples for research.
Discussion
Esophageal carcinosarcoma is a rare kind of malignancy with controversial characteristics and poor prognosis. Recent studies discover that esophageal carcinosarcoma appeared to be comprised of different origins of epithelial and mesenchymal tumor cells [5]. The fibroblast was evident in this case compared with ESCC (Figure 1b), however the morphology of spindles-shaped cells in EC is difficult to distinguish with spindle cell carcinoma. Therefore, immunohistochemistry is necessarily indicative for the origin of spindle cell [6]. As seen in this case, the squamous cancer cells are positive for CK, nevertheless the spindle cells are positive for vimentin. Surgery with extended lymphadenectomy for esophageal carcinosarcoma is the standard treatment, but chemotherapy may be a good choice for local control for patients who cannot undergo surgical resection. Actually, esophageal carcinosarcoma is locally aggressive and can recur quickly after resection with poor prognosis. In a 2006 review, only 13% of resected patients were reported to be alive “free of disease” at 2 years after surgery [7]. And the rate of resected patients alive “free of disease” or dead from unrelated causes dropped from about 60% at 3 years after surgery to about 15% at 5 years [8]. Noticeably, in this case, the patient had no lymphatic metastasis before the curative resection, while the tumor recurred quickly and he lived just 9 months after surgery with systemic metastases.
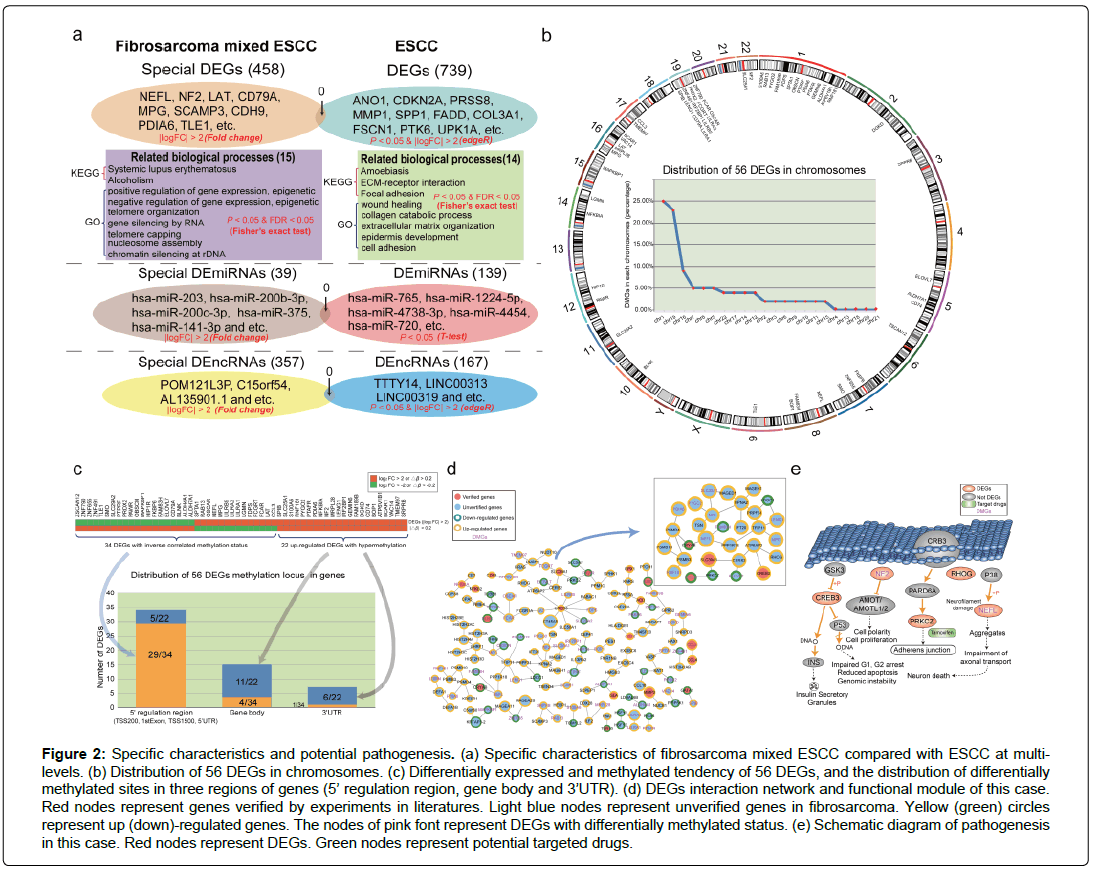
Given the controversial characteristics and the rarity of esophageal carcinosarcoma, its pathogenesis is obscure. Therefore, exploring the potential mechanism and revealing the abnormal molecules in this complex malignancy are conducive to elucidating the pathogenesis and providing personalized treatment strategies. With the development of bioinformatics, analyzing high-throughput data of rare diseases could contribute to identifying crucial abnormal genes and dysfunction biological pathways. Here, we explore the molecular characteristics of this case based on the corresponding multi-omics data. We found that most telomere and epigenetic biological processes were disordered in this case based on differentially expressed genes (DEGs). Interestingly, the abnormal biological processes of ESCC were completely different with that of this case (mainly focused on well-known cancer related functions, such as focal adhesion and epidermis development (Figure 2a)), which indicate that carcinosarcoma and squamous cell carcinoma coexisting in the esophagus has special pathogenesis compared with ESCC. The DEGs and aberrant biological pathways were provided in Supplementary Table 1 and Supplementary Table 2, respectively. Remarkably, aberrant non-coding RNAs (provided in Supplementary Table 3 and Supplementary Table 4), particularly differentially expressed lncRNAs (DElncRNAs) were distinct in two diseases, which imply that the abnormal regulation of non-coding RNAs might lead to the specific characteristics of this case (Figure 2a). Furthermore, DNA methylation status of this rare disease was significant changed compared with normal sample. 56 DEGs were identified with aberrant DNA methylation status (Figure 2b). We found that most aberrant DNA methylation status occurred in 5’ regulation region and influenced the expression of the corresponding DEGs, while occurred in gene body or 3’UTR did not disturb genes’ expression (Figure 2c). These results demonstrate that multi-level disorder of this rare disease might lead to the further deterioration. Clarifying the interaction (Figure 2d) of these aberrant molecules can be conductive to confirming the pathogenesis of this case. We identified the abnormal expressed gene, CREB3, which affected the processes of insulin secretory granules and cell cycle by secreting DNA in fibrosarcoma (Figure 2e). In addition, NF2, a DEG with differentially methylated status, inhibited the expression of Angiomotin (AMOT) and influenced cell polarity and proliferation (Figure 2e). In cyctoplasm, the dysregulated PRKCZ and NEFL disturbed adherens junction and neuron death processes (Figure 2e). These abnormal processes might be the potential pathogenesis of this case.
Figure 2: Specific characteristics and potential pathogenesis. (a) Specific characteristics of fibrosarcoma mixed ESCC compared with ESCC at multilevels. (b) Distribution of 56 DEGs in chromosomes. (c) Differentially expressed and methylated tendency of 56 DEGs, and the distribution of differentially methylated sites in three regions of genes (5’ regulation region, gene body and 3’UTR). (d) DEGs interaction network and functional module of this case. Red nodes represent genes verified by experiments in literatures. Light blue nodes represent unverified genes in fibrosarcoma. Yellow (green) circles represent up (down)-regulated genes. The nodes of pink font represent DEGs with differentially methylated status. (e) Schematic diagram of pathogenesis in this case. Red nodes represent DEGs. Green nodes represent potential targeted drugs.
Noticeably, it is generally treated by operation, radiotherapy and chemotherapy according to the common protocols for esophageal tumors. Recent study find that the tumor was remarkable reduction only treated with neoadjuvant radiotherapy (40 Gy) before surgery in esophageal carcinosarcoma and radiotherapy alone might be a suitable neoadjuvant therapy for esophageal carcinosarcomas (Ogasawara et al, 2014). In this case, the patient underwent radiotherapy with a dose of 50 Gy (Partially increased to 64-66 Gy), and concurrent chemotherapy with Tegafur/Uracil after surgery, while the tumor still recurred and the patient died in a short period of time after surgery. Tegafur/Uracil is the chemotherapy drug of this case, which is approved in FDA and indicated for the first line treatment of metastatic colorectal cancer with concomitant administration of calcium folinate [9]. In tumors, Tegafur/Uracil targets DPYP and TYMS to decreased thymidine synthesis, DNA synthesis, disrupted RNA function and tumor cell cytotoxicity [9]. However, in this case, the expression of DPYP (log |FC|=1.77) and TYMS (log |FC|=1.46) shows no significant difference between tumor and matched normal samples. That is, these two genes are negative in this case. These phenomena explained why the effect of chemotherapy (Tegafur/Uracil) was poor after surgery. To predict potential effective drugs of this case, we collected the drugs targeted 56 DEGs and inferred them as the potential drugs. For example, PRKCZ (log |FC|>2) is aberrant in this case and tamoxifen is a clinical drug to prevent and treat breast cancer in women by targeting PRKCZ [9]. We inferred that tamoxifen might target PRKCZ and prevent the deterioration of this case. In addition, the patient had no lymphatic metastasis before the curative resection, while he lived just 9 months after surgery with systemic metastases. Metastasis involves a complex series of steps and seems to be mediated by soluble signal molecules, such as chemokines and transforming growth factor [10-12]. By analyzing the corresponding multi-omics data of this case, we discovered several well-known chemokines (CCL3, CCL4, CCL23 and CCL24) were dysfunction, which might imply that the tumor would migrate after surgery. These results suggest that integrating multi-omics data in rare diseases could provide potential therapeutic strategies to clinical research and accelerate the development of precision medicine. The corresponding multi-omics data of this case was uploaded to GEO database (Accession number: GSE112083, GSE112030 and GSE112012). The detailed bioinformatics analysis was listed in Supplementary information file.
Conclusion
Carcinosarcoma and squamous cell carcinoma coexisting in the esophagus is an uncommon malignancy that presents poor prognosis and high recurrence rate. Both carcinomatous and sarcomatous components are seen at histological level in this case, but the pathogenesis is unclear. In this report, we obtained the sequencing data (RNA-seq, DNA methylation), and identified the differentially expressed molecules, such as DEGs and DElncRNAs. Then we calculated the correlation (Pearson) of these abnormal molecules and explored the potential mechanism of this rare case both in transcriptional and post-transcriptional level. Integrating multiomics data will be a comprehensive approach to screen molecular characteristics, define the connection between patient phenomes and molecules, and elucidate the pathogenesis of diseases in individual cases, which could provide novel therapeutic strategies in the development of precision medicine.
References
- Pennathur A, Gibson MK, Jobe BA, Luketich JD (2013) Oesophageal carcinoma. Lancet 381: 400-412.
- Cavallin F, Scarpa M, Alfieri R, Cagol M, Ruol A, et al. (2014) Esophageal carcinosarcoma: Management and prognosis at a single Italian series. Anticancer Res 34: 7455-7459.
- Yoshimoto T, Kobayashi S, Kanetaka K, Kobayashi K, Nagata Y, et al. (2018) Preoperative chemotherapy with docetaxel, cisplatin, and 5-fluorouracil for locally advanced esophageal carcinosarcoma: A case report and review of the literature. Surg Case Rep 4: 18.
- Kumar-Sinha C, Chinnaiyan AM (2018) Precision oncology in the age of integrative genomics. Nat Biotechnol 36: 46-60.
- Miyauchi J, Ogura M, Sato M, Matsui J (2018) Esophageal carcinosarcoma comprised of minimally invasive squamous cell carcinoma and undifferentiated pleomorphic sarcoma: A collision cancer? Pathol Int .
- Akagi I, Miyashita M, Makino H, Nomura T, Ohkawa K, et al. (2008) So-called carcinosarcoma of the esophagus: Report of a case. J Nippon Med Sch 75: 171-174.
- Sanada Y, Hihara J, Yoshida K, Yamaguchi Y (2006) Esophageal carcinosarcoma with intramural metastasis. Dis Esophagus 19: 119-131.
- Iascone C, Barreca M (1999) Carcinosarcoma and pseudosarcoma of the esophagus: Two names, one disease-comprehensive review of the literature. World J Surg 23: 153-157.
- Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, et al. (2018) DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res 46: D1074-D1082.
- Nguyen DX, Massague J (2007) Genetic determinants of cancer metastasis. Nat Rev Genet 8: 341-352.
- Zlotnik A, Burkhardt AM, Homey B (2011) Homeostatic chemokine receptors and organ-specific metastasis. Nat Rev Immunol 11: 597-606.
- Drabsch Y, ten Dijke P (2011) TGF-beta signaling in breast cancer cell invasion and bone metastasis. J Mammary Gland Biol Neoplasia 16: 97-108.