Research Article, J Appl Bioinformat Computat Biol S Vol: 0 Issue: 0
Screening and Identification of Antiviral Drugs from Drug Bank Database Targeting SARSCov- 2 Non-Structural Proteins (NSP): A Virtual Screening and Molecular Docking Study
Alam MM*, Shill DK, Jahan S, Alam M, Hossain ME, Rahman M and Rahman MZ
Virology Laboratory, International Centre for Diarrhoeal Disease Research, Mohakhali, Dhaka 1212, Bangladesh
*Corresponding Author: Alam MM
Virology Laboratory, International Centre for Diarrhoeal Disease Research
Mohakhali, Dhaka-1212, Bangladesh
E-mail: mamun.alam@icddrb.org
Received: October 13, 2021 Accepted: October 27, 2021 Published: November 03, 2021
Citation:Alam MM, Shill DK, Jahan S, Alam M, Hossain ME, et al. (2021) Screening and Identification of Antiviral Drugs from Drug Bank Database Targeting SARS-Cov-2 Non-Structural Proteins (NSP): A Virtual Screening and Molecular Docking Study. J Appl Bioinforma Comput Biol S5.
Abstract
The ongoing pandemic of Severe Acute Respiratory Syndrome Corona Virus 2 (SARS-CoV-2) is a positive-sense RNA (ss) virus. Mutational change is a typical episode resulting in their emerging regional variants not responding to the vaccine equally. Therefore, along with vaccines, antiviral drugs targeting non-structural proteins (nsp) can be a remedy to provide maximum protection. Moreover, in immune compromised individuals, antiviral drugs can be the only reliable choice to tackle these types of infections. Here, nonstructural proteins (nsp) named RNA dependent RNA polymerase (RdRp), Helicase (NSP13), and Papain-like Protease (NSP3) were selected as the target for drugs that can provide protection irrespective of mutational variants. Validated structures of these three essential proteins used to search anti-SARS-CoV-2 drugs (in silico). DrugBank database suggests eight drugs, including Remdesivir General, that can react against these three nonstructural proteins. Molecular docking with AutoDock vina tools identified two potential drug-like components such as Nalpha- [(benzyloxy) carbonyl]-N-[(1R)-4-hydroxy-1-methyl-2-oxobutyl]-Lphenylalaninamide (DB08732), and S- [5-(trifluoromethyl L)-4H- 1,2,4-triazol-3-YL] 5-(phenylethyl) furan-2-carbothiote (DB07743) showed significant binding energy against these proteins such as -54.39 KJ/mole and -52.3 KJ/mole, respectively. Also, the ADMET profile showed that DB08732 and DB07743 have no carcinogenicity. In addition, no ortholog of these three proteins was found in the human body, supporting their antiviral drug-like potential for treating SARS-CoV-2. Reflects that DB08732 and DB07743 might be promising candidates for therapeutic intervention to block SARSCoV- 2 replication to the host cell along with vaccines
Keywords: SARS-Cov-2; Antiviral Drug; Rdrp; Helicase; Plpro; Molecular Docking
Keywords
SARS Cov 2; Antiviral Drug; Rdrp; Helicase; Plpro; Molecular Docking
Abbreviation
ADMET ( Distribution, Metabolism, Excretion, and Toxicity); PROCHECK (Program to Check the Stereochemical Quality of Protein Structures); BLAST (Basic Local Alignment Search Tool); PROSA (Protein Structure Analysis).
Introduction
In December 2019, a newly emerged human coronavirus became the focus of global concern after the dramatic and widespread outbreak of mystic pneumonia-like respiratory illness in the Wuhan region of Hubai, the province in China. On February 11, 2020, The International Virus Classification Commission (ICTV) designated the virus as Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2). Later the disease caused by SARS-CoV-2 was named COVID-19 by WHO. In March 2020, COVID-19 was declared a Public Health 1 Emergency of International Concern (PHEIC), consequently pandemic by WHO [1].
Based on genomic alikeness and evidence from phylogenetic analysis, SARS-CoV-2 is considered a member of Betacoronaviruses, including SARS-CoV, MERS-CoV [2]. SARS-CoV-2 showed more than 80% sequence similarity with SARS-CoV and 50% with the MERS-CoV [3]. Also, these viruses might use the same receptor because of the amino acid sequence resemblance of the tentative receptor-binding domain [4].
The SARS-CoV-2 is an enveloped, positive-sense, single-stranded RNA virus with a genome size of 29.7 kb [5]. Upon internalization into the cell, genomic RNA is composed of six open reading frames (ORFs). The first ORF (ORF1a/b) is vital, occupying two-thirds of the genome. ORF1a/b was used as a template for translating two polyproteins, pp1a and pp1ab, which encodes the production of 16 non-structural proteins (nsp) [6]. Some of these nsp encode proteins with essential functions, such as Papain-like protease, PLpro (nsp3), Chymotrypsin-like protease, CLpro (nsp5), Helicase (nsp13), and RNA dependent RNA polymerase, RdRP (nsp12). CLpro & PLpro cleave the replicase polyprotein in a sequence-specific manner to produce 15 to 16 non-structural proteins (nsp). In contrast, Helicase (nsp13) unwinds duplex oligonucleotides in an NTP-dependent way. RdRp plays a critical role in the viral infection cycle by forming a replication-transcription complex [7].
The rapidly growing cases of COVID-19 lead to the rapid development of new therapeutics and vaccines against SARS-CoV-2. There are multiple phases of vaccine development, including an initial design stage, preclinical studies, and phases 1–3 of clinical trial testing. However, researchers, governments, pharmaceutical companies, and regulatory bodies are tackling the spread of the pandemic virus by coordinating an unprecedented overhaul of the vaccine development process. Vaccines are very effective on stable viruses (Hepatitis B virus, Adenovirus) but are limited in treating already infected patients. Therefore, it becomes difficult to successfully utilize them against rapidly mutating viruses, such as influenza (the vaccine needed to update every year), HIV & HCV [8].
As a possible rapid therapeutics approach against rapidly spreading emerging infections like SARS-CoV-2, Drug repurposing has emerged as a substitution of a vaccine. Recently, several studies using clinical drugs against SARS-CoV-2 have been proposed by scientists worldwide.
For a practical therapeutic approach, the active site of RdRp as the most conserved and accessible region constitutes a significant target for inhibition of viral replication. In addition, to block the production of non-structural viral components, followed by a hamper of virus replication, the papain-like protease and Helicase enzyme leads to an attractive drug target.
We used bioinformatics tools to identify SARS-CoV-2 encoded specified proteins and retrieve their sequences for mutational analyses. In addition, performed homology modeling to build targeted protein structures, such as papain-like protease (PLpro), RNA-dependent RNA polymerase (RdRp), Helicase, etc. The main objective was to screen drugs (experimental or approved) from the drug bank targeting non-structural proteins through in silico analysis. Thus, our study screens various components that may inhibit SARS-CoV-2 and provide a good starting point for antiviral therapies as an alternative or coadjuvant vaccination strategies. Furthermore, subsequent validation of antiviral effects in vitro and in vivo will provide helpful information for the clinical treatment of novel coronavirus.
Materials and Methods
Identification of the Sole Proteins Involved in the Life Cycle of SARS-CoV-2
We identified the sole proteins (RdRp, Helicase, and PLpro) involved in the life cycle of SARS-CoV-2 through literature analysis and explored their antiviral drugs [9,10].
Mutation Analysis of sole proteins among Regional Variants
The amino acid sequences of identified proteins from different regional variants, for instance, Indian variant (QUE93972), South African Variant (QUA12568.1.), Brazilian variant (QMB22609.1), U.K. variant (QOS14143.1), emerging variants of Bangladesh (QKV26694.1, QWT51802.1) and Wuhan strain (YP_009725308) as a reference, were retrieved from NCBI database. Then, the ClustalW method of Multiple Sequence Alignment (MSA) was used with the bootstrap value of 1000 in conjunction with BioEdit software [11].
Protein Modeling
We performed homology modeling of RdRp (NCBI ID QKV26694.1), Helicase (NCBI ID QKV26694.1), and Papain Like protease (PLpro) (NCBI ID QKV26694.1) using the SWISS- MODEL database [12]. We used 7cyq.1.A (RdRp), 5rl6.2.A (Helicase), 6wuu.2.A (L-protease) template which determines the crystal 3D structure with the sequence identity around 99.89 %, 100 %, 99 %, respectively. We also used PyMol for structure visualization [13]. For protein 3D structure quality assessment, PROCHECK [14] was used and validated with SAVES-Verify3D [15], PROSA- Web Server [16], Ramachandra Plotting. Finally, we used ExPASy: Protpram tools to determine physicochemical properties [17]. The atomic charge calculator was used to determine the charge of these three proteins [18].
Retrieval of Drug-Like Components from Drug Bank
Protein sequences were used to model, identify drug-like components, and determine pharmacological function through DrugBank Database (https://go.drugbank.com) [19]. The sequence of drug-like components (experimental or approved) that can interact and interfere with proteins (sdf or.pdb) were retrieved. Later the sequences were used to determine binding efficacy analysis with respective proteins. And, ADMET profiling was determined by admetSAR [20].
Molecular Docking
The AutoDock Vina 1.1.5 tool was used [21], used to conduct target protein interactions with ligand drug components. We set proteins dehydration, protonation, and specific grid box with grid point spacing 1 Å within proteins. Then, formatted the protein and ligand sequences with Open Babel [22]. Molecular docking was performed with an exhaustiveness value of eight. Based on protein-ligand interactions, binding energy (Kcal/mole) was released. We used Pymol 3D viewer as well as BIOVIA Discovery Studio Visualizer software to visualize protein-ligand three-dimensional interactions.After performing molecular docking analysis, interacting amino acids in the binding pocket of protein with respective type of interactions were also visualized with BIOVIA Discovery Studio Visualizer software [23]. The X, Y, Z coordinate of the binding site region of each targeted protein were determined. And, the type of charges of proteins and ligands were also determined.
Broad Spectrum Homology analysis with Homo sapiens
Protein sequences were subjected to pBLAST tools of NCBI [24] to determine whether pathogenic viruses contain these proteins or not. Additionally, whether Homo sapiens has an ortholog any of this protein or not (E value 0.0001).
Results
Sole Proteins Involved in the Multiplication of SARS-CoV-2
SARS-CoV-2 has three unique non-structural proteins named RdRp, Helicase, and Papain Like-protease (PLpro). RdRp plays a crucial role in replication and can be an ideal target for antiviral drugs. Whereas, Helicase [25] and Papain Like-protease (PLpro) [26] play a significant role in virus propagation. All three proteins play a vital role in genome replicating and propagating in the host cell [27]. We know that mutation quickly takes place in the structural proteins such as a spike or envelope proteins. Therefore, we opt to identify drugs irrespective of mutational resistance as they act on non-structural proteins activity. Furthermore, we tried to find out the level of cure they could provide. Therefore, antiviral drugs that can inhibit nsp’s function can inhibit virus replication and propagation.
Mutational Analysis
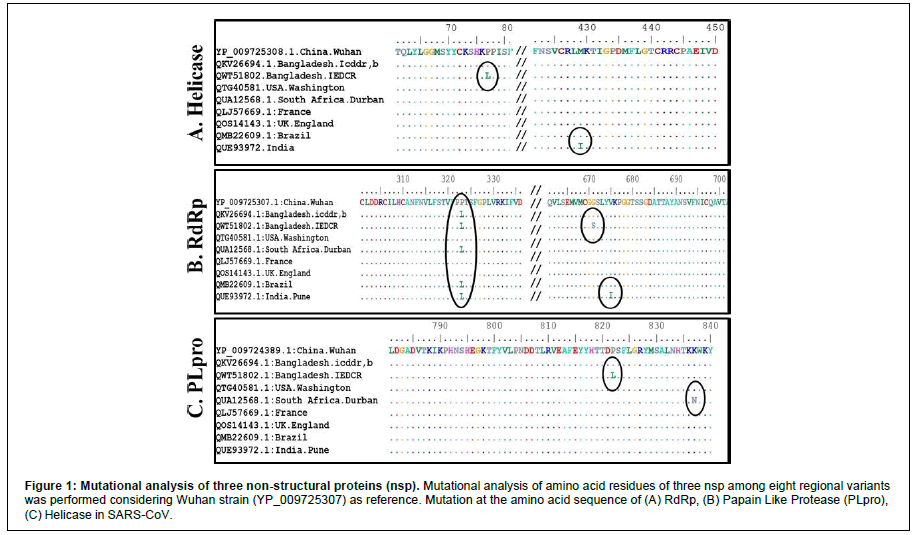
We aligned sequences of proteins (RdRp, Heliase, PLpro) from eight regional variants by ClustalW (Figure 1). In Helicase (Figure 1A), amino acids mutation (Proline to Leucine) at position number 77 in emerging Indian Delta variant isolated in Bangladesh (QWT51802.1), and another mutation (Methionine to Isoleucine) at 429 position of Indian variant isolated in India was found compared with Wuhan strain. In RdRp (Figure 1B), at 323 position Bangladesh (icddr,b; IEDCR), South African (QUA12568.1), Indian, and Brazilian variant (QMB22609.1) have similar Proline to Leucine mutation. Whereas, Indian variant isolated from Bangladesh (IEDCR) showed mutation at position 671 (Glycine to Serine) and the Indian variant in India at position 675 (Valine to Isoleucine). However, Papain Like Protease (Figure 1C) has only two mutations; one in the Indian variant isolated from Bangladesh (IEDCR) at position 822 (Proline to Isoleucine) and South African variant at position 837 (Lysine to Asparagine).
Figure 1: Mutational analysis of three non structural proteins (nsp). Mutational analysis of amino acid residues of three nsp among eight regional variants was performed considering Wuhan strain (YP_009725307) as reference. Mutation at the amino acid sequence of (A) (B) Papain Like Protease ( (C) Helicase in SARS CoV.
Protein Modeling and Validation
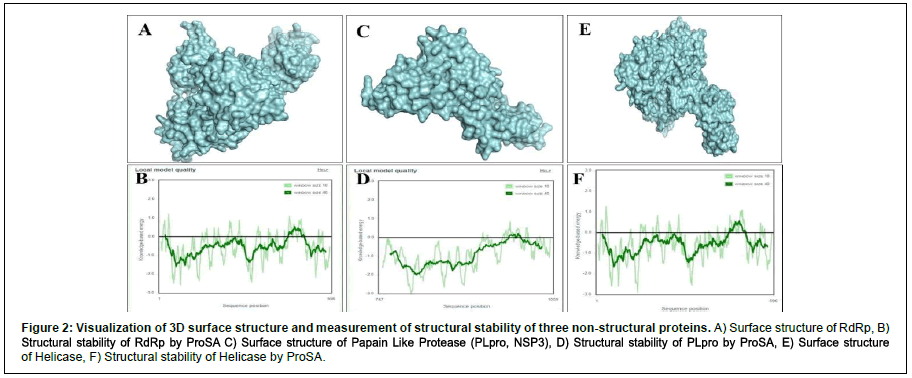
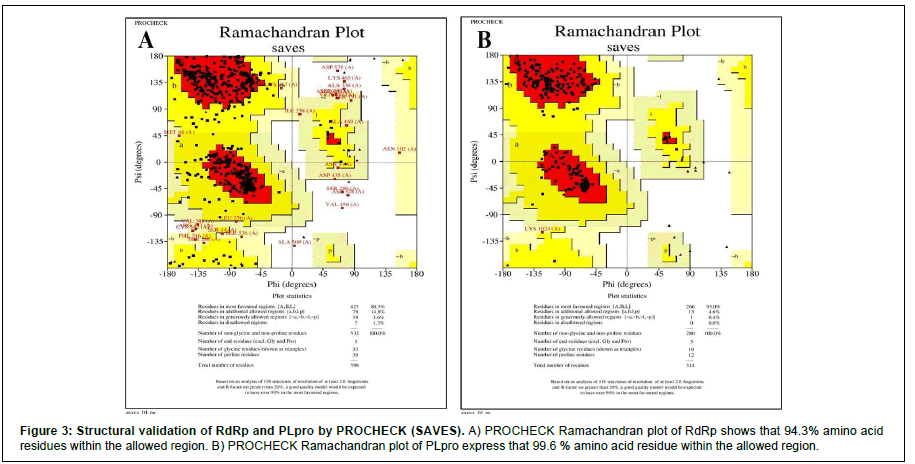
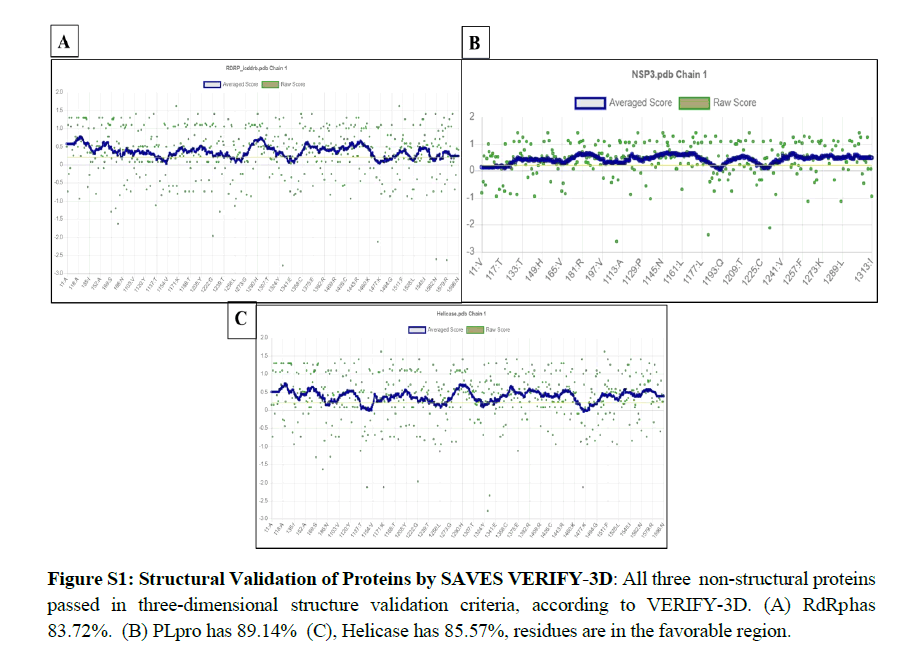
We performed protein modeling based on the homology modeling algorithm using the SWISS-MODEL database. First, we visualized the 3D model using.pdb format with PyMOL software (Figure 2). Then, the 3D structure was verified with PROCHECK, SAVES-Verify 3D, PROSA-Web, and Ramachandra Plotting. Helicase and PL protease have 85.57% and 89.14% residues (Table S1), respectively, in the favorable area. Whereas for RdRp, it is 83.72%. Ramachandran Plotting showed RdRp (Figure 3A), Helicase, and PLpro (Figure. 3B) have 95.1%, 99%, and 99.6% amino acids residues resided allowed region (Figure 3), respectively including 80.3%, 76.3%, and 95% in the most favored area. Finally, the PROSA-web service determined the overall stability of the protein 3D structure. All three showed stable structure, having Z values -8.9, -8.89, and -8.85 for RdRp (Figure. 2B), PL protease (Figure 2D), and Helicase (Figure 2F), respectively. RMSD (Root Mean Square Distance) values of these three protein models with their respective templates are 1.776 Å (RdRp), 1.640 Å (Helicase) and 0.822 Å (PLpro). RMSD values less than 2 Å are considered as validated structures. Physiochemical properties with charge are enlisted in Table 1.
Figure 2: Visualization of 3D surface structure and measurement of structural stability of three non structural proteins. A) Surface structure of RdRp, B) Structural stability of RdRp by ProSA C) Surface structure of Papain Like Protease (PLpro, NSP3), D) Structural stability of PLpro by ProSA, E) Surface structure of Helicase, F) Structural stability of Helicase by ProSA.
| Physicochemical Properties | Helicase | RdRp | PL Protease (NSP3) |
|---|---|---|---|
| Number of amino acids | 601 | 932 | 1945 |
| Molecular weight | 66855 | 106676.28 | 217253 |
| Theoritical pI | 8.66 | 6.14 | 5.56 |
| Total number of negatively charged residues (Glu + Asp) | 52 | 106 | 222 |
| Total number of Positively charged residues (Lys + Arg) | 64 | 94 | 185 |
| Extinction coefficient assuming all pairs of Cys residues from cysteines M-1 cm-1 | 68785 | 137670 | 243675 |
| Extinction coefficient assuming all pairs of Cys residues reduced� M-1 cm-1 | 67160 | 135920 | 240550 |
| Instability index | 33.31 | 8.12 | 36.56 |
| Aliphatic index | 84.49 | 78.85 | 86.22 |
| Grand average of hydropathicity (GRAVY) | -0.096 | -0.218 | -0.175 |
| Physiological charge | 0 | 0 | 0 |
� Note pI: Isoelectric point, RdRp: RNA dependent RNA polymerase, NSP: Non-structural Protein).
Table 1: �Physicochemical Properties of Three Non-Structural Proteins (Nsp).
Inhibition of Sole Proteins by Drugbank Database Suggest- ed Drugs
Sequence-based in silico analysis of DrugBank database suggested eight similar and one (only against PL-protease) chemical compounds (Table 2) that as well as the activity of RdRp, Helicase, and Papain Like-protease (PLpro) proteins. Among these experimental drugs, only Remdesivir General was found to have Food and Drug Administration (FDA) approval but investigational to treat COVID-19 patients [28].
| Name of the drug | Drug Bank ID | Physiological Charge | Docking score/ Binding Energy (Kcal/mole) with Specified Protein | ||
|---|---|---|---|---|---|
| RNA Dependent RNA Polymerase (RdRp) | Helicase | Papain Like Protease (PLpro) | |||
| Benzyl (2-oxopropyl) carbamate | DB07293 | 0 | -8.7 | -7.7 | -8.1 |
| 2- [(2,4-Dichloro-5-methyl phenyl) sulfonyl]-1,3-dinitro-5-(trifluoromethyl)benzene | DB07620 | 0 | -13.1 | -13.1 | -13.1 |
| S- [5-( trifluoromethyl L)-4H-1,2,4-triazol-3-YL] 5-(phenylethyl) furan-2-carbothiote | DB07743 | -1 | -12.5 | -13.4 | -11.4 |
| 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl] benzamide | DB08656 | 0 | -11.1 | -10.5 | -9.4 |
| Nalpha-[(benzyloxy) carbonyl]-N-[(1R)-4-hydroxy-1-methyl-2-oxobutyl]-L-phenylalaninamide | DB08732 | 0 | -13 | -12.5 | -11.8 |
| 4-(Dimethylamino)benzoic acid | DB08748 | -1 | -6.9 | -6.9 | -7.7 |
| Remdesivir General | DB14761 | 0 | -16 | -15.3 | -13.3 |
| GS-441524 | DB15686 | 0 | -9.3 | -11.2 | -10.1 |
| 2-(N-morpholino)ethanesulfonic acid | DB03814 | -1 | N/A | N/A | -6.3 |
Table 2: Molecular Docking Score or Binding Energy of Nine Different Drug-Like Components with their Protein.
Molecular Docking
Finally, nine drugs were subjected to binding Analysis using the molecular docking method. We used AutoDock tools-1.1.5 to determine drugs’ binding affinity and efficiency with specified protein by observing their binding energy with proteins (Table 2) [21].
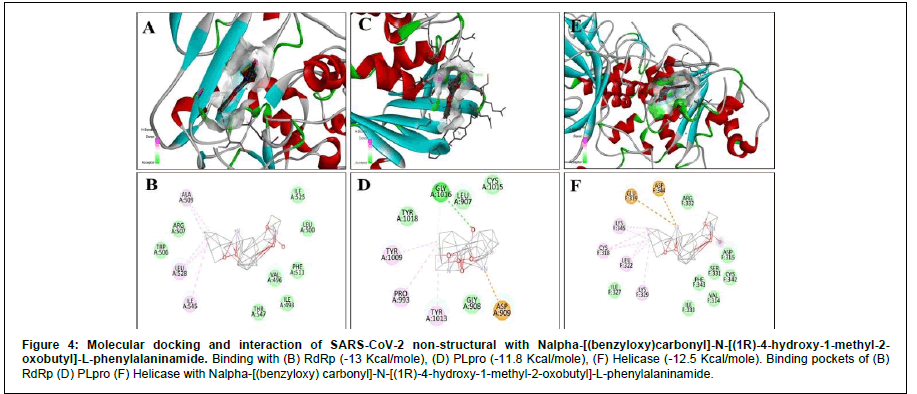
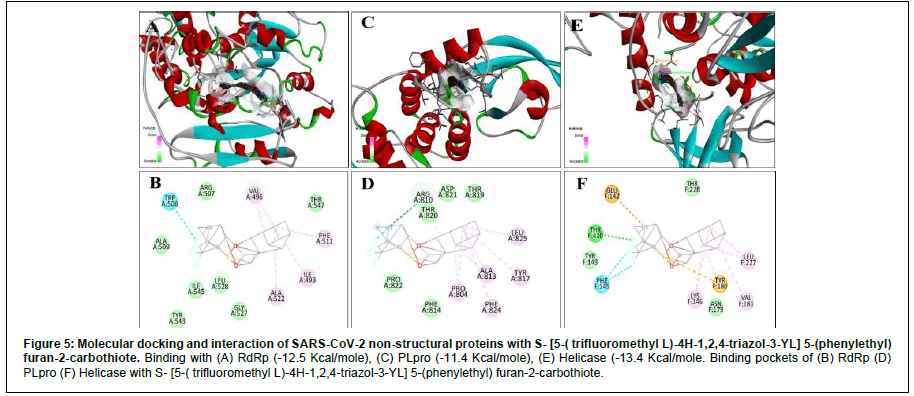
For RdRp, apart from Remdesivir General, the energy expressed in negative signs denotes that these reactions are exothermic (Table 2). Exergonic energy is more than eight Kcal/mole recommended as effective against the specific proteins (Table 2) [29]. We found 2- [(2,4-Dichloro-5-methyl phenyl) sulfonyl]-1,3-dinitro-5- (trifluoromethyl) benzene, Nalpha-[(benzyloxy) carbonyl]-N-[(1R)- 4-hydroxy-1-methyl-2-oxobutyl]-L-phenylalaninamide and S- [5-( trifluoromethyl L)-4H-1,2,4-triazol-3-YL] 5-(phenylethyl) furan-2- carbothiote showed binding energy – 8.7 Kcal/mole, - 13 Kcal/mole (Figure 4) and – 12.5 Kcal/mole (Figure 5), respectively (Table 2). Interacting amino acids (5) with S- [5-( trifluoromethyl L)-4H-1,2,4- triazol-3-YL] 5-(phenylethyl) furan-2-carbothiote showed Halogen bond as well as strong Alkly bonds (Table 3). Moreover, in case of Nalpha-[(benzyloxy) carbonyl]-N-[(1R)-4-hydroxy-1-methyl-2- oxobutyl]-L-phenylalaninamide, we found 3 interacting amino acids with strong Alkyl bonds (Table 3).
| Enzyme | Drug Name | Drug Bank ID | Associated Amino Acid (Position) | Types of bonding |
|---|---|---|---|---|
| RdRp | S- [5-( trifluoromethyl L)-4H-1,2,4-triazol-3-YL] 5-(phenylethyl) furan-2-carbothiote | DB07743 | W (506) | Halogen (Fluorine) |
| V (496), F (511), I (493), A (522) | Alkyl | |||
| Nalpha-[(benzyloxy) carbonyl]-N-[(1R)-4-hydroxy-1-methyl-2-oxobutyl]-L-phenylalaninamide | DB08732 | A (509) L (528), I (545) | Alkyl | |
| Helicase | S- [5-( trifluoromethyl L)-4H-1,2,4-triazol-3-YL] 5-(phenylethyl) furan-2-carbothiote | DB07743 | T (410) | Conventional Hydrogen bond |
| L (227), K (146), V (181) | Alkyl | |||
| F (145) | Halogen (Fluorine) | |||
| E (142) | Attractive Charge | |||
| Y (180) | Pi-Sulfur | |||
| Nalpha-[(benzyloxy) carbonyl]-N-[(1R)-4-hydroxy-1-methyl-2-oxobutyl]-L-phenylalaninamide | DB08732 | E (319), D (344) | Attractive Charge | |
| K (345, 329), C (318), L (322) | Alkyl | |||
| PLPro | S- [5-( trifluoromethyl L)-4H-1,2,4-triazol-3-YL] 5-(phenylethyl) furan-2-carbothiote | DB07743 | L (825), A (813), P (804), F (804), Y (817) | Alkyl |
| R (810) | Halogen (Fluorine) | |||
| R (810) | Unfavorable Positive-Positive Charge | |||
| T (820) | Van Der Walls | |||
| Nalpha-[(benzyloxy) carbonyl]-N-[(1R)-4-hydroxy-1-methyl-2-oxobutyl]-L-phenylalaninamide | DB08732 | G (1016) | Conventional Hydrogen bond | |
| N (909) | Attractive Charge | |||
| T (1009,1013), P (993) | Alkyl |
Table 3: Name interacting amino acids within the protein and their interactions with drug-like components.
For Helicase (NSP13), the S- [5-( trifluoromethyl L)-4H- 1,2,4-triazol-3-YL] 5-(phenylethyl) furan-2-carbothiote showed the most effective binding (Figure 5) that is – 13.4 Kcal/mole. In this interaction, seven amino acids are involved in a pool of interactions (Conventional Halogen, Alkyl, Pi-Sulfur and attractive charge bonding). Although, 2- [(2,4-Dichloro-5-methyl phenyl) sulfonyl]-1,3-dinitro-5-(trifluoromethyl) benzene (- 13.1 Kcal/mole) and Nalpha-[(benzyloxy) carbonyl]-N-[(1R)-4-hydroxy-1-methyl- 2-oxobutyl]-L-phenylalaninamide(-12.5 Kcal/mole) also showed impressive binding response (Figure 4). Here, five amino acids are involved in the interaction with Nalpha-[(benzyloxy) carbonyl]-N- [(1R)-4-hydroxy-1-methyl-2-oxobutyl]-L-phenylalaninamide with two types of bonds (Table 3).
Figure 5: Molecular docking and interaction of SARS-CoV-2 non-structural with Nalpha-[(benzyloxy)carbonyl]-N-[(1R)-4-hydroxy-1-methyl-2-oxobutyl]-L-phenylalaninamide. Binding with (B) RdRp (-13 Kcal/mole), (D) PLpro (-11.8 Kcal/mole), (F) Helicase (-12.5 Kcal/mole). Binding pockets of (B) RdRp (D) PLpro (F) Helicase with Nalpha-[(benzyloxy) carbonyl]-N-[(1R)-4-hydroxy-1-methyl-2-oxobutyl]-L-phenylalaninamide.
Figure 6: Molecular docking and interaction of SARS-CoV-2 non-structural proteins with S- [5-( trifluoromethyl L)-4H-1,2,4-triazol-3-YL] 5-(phenylethyl) furan-2-carbothiote. Binding with (A) RdRp (-12.5 Kcal/mole), (C) PLpro (-11.4 Kcal/mole), (E) Helicase (-13.4 Kcal/mole. Binding pockets of (B) RdRp (D) PLpro (F) Helicase with S- [5-( trifluoromethyl L)-4H-1,2,4-triazol-3-YL] 5-(phenylethyl) furan-2-carbothiote.
In case of PL protease, Nalpha-[(benzyloxy) carbonyl]-N-[(1R)- 4-hydroxy-1-methyl-2-oxobutyl]-L-phenylalaninamide showed – 11.8 Kcal/mole (Figure 4), 2- [(2,4-Dichloro-5-methyl phenyl) sulfonyl]-1,3-dinitro-5-(trifluoromethyl) benzene showed – 13.1 Kcal/mole, and S- [5-( trifluoromethyl L)-4H-1,2,4-triazol-3-YL] 5-(phenylethyl) furan-2-carbothiote has – 11.4 Kcal/mole (Figure 5) as binding energy (Table 2). X, Y, Z coordinate of the binding site region of each targeted protein with Nalpha-[(benzyloxy) carbonyl]- N-[(1R)-4-hydroxy-1-methyl-2-oxobutyl]-L-phenylalaninamide and S- [5-( trifluoromethyl L)-4H-1,2,4-triazol-3-YL] 5-(phenylethyl) furan-2-carbothiote were enlisted (Table 4). In case of Nalpha- [(benzyloxy) carbonyl]-N-[(1R)-4-hydroxy-1-methyl-2-oxobutyl]- L-phenylalaninamide, four amino acids are involved in three types of interactions (Conventional Hydrogen, attractive charge and Alkyl) (Table 3). Furthermore, eight amino acids are involved in the interaction with S- [5-( trifluoromethyl L)-4H-1,2,4-triazol-3-YL] 5-(phenylethyl) furan-2-carbothiote along with four types of strong bonds (Van Der Wales, Alkyl, Halogen and Unvaourable positive- positive charge) (Table 3).
| Protein | Ligand | X coordination | Y coordination | Z coordination |
|---|---|---|---|---|
| RdRp | DB07743 | 391.237 | -11.811 | 87.190 |
| DB08732 | 394.080 | -15.572 | 86.892 | |
| Helicase | DB07743 | 222.672 | 185.026 | 157.543 |
| DB08732 | 209.463 | 211.549 | 155.121 | |
| PLpro | DB07743 | -23.596 | 23.779 | -41.256 |
| DB08732 | -39.372 | -3.713 | -39.372 |
Table 4: X, Y, Z coordination of binding site of targeted protein with specified drug-like components.
Properties and Pharmacological Mode of Action of these Drug Compounds
Remdesivir General and GS-441524 were already involved in the inhibitory action; the rest have unknown pharmacological mode of action (Figure S1). According to the admetSAR 2.0, Nalpha- [(benzyloxy) carbonyl]-N-[(1R)-4-hydroxy-1-methyl-2-oxobutyl]- L-phenylalaninamide, S- [5-( trifluoromethyl L)-4H-1,2,4-triazol- 3-YL] 5-(phenylethyl) furan-2-carbothiote and GS-441524 showed good drug-like properties to consider as a lead component for the drug discovery process, with no carcinogenic or eye corrosion or irritation effect. However, 2- [(2,4-Dichloro-5-methyl phenyl) sulfonyl]-1,3-dinitro-5-(trifluoromethyl)benzene has a bit of binary carcinogenic and eye corrosion effect. Physiological charges of these drug-like components were determined from ChemAxon (http//: chemaxon.com) database (Table 2). According to this database only DB07743, DB08748 and DB03814 are in negative (-1) charge and rest of them are in neutral charge (Figure 4).
Broad Spectrum Analysis
We conducted pBLAST analysis to determine whether the human genome contains these types of proteins or not. If the human genome contains protein like this, it will be a great trouble administering this drug to the human body. However, after pBLAST analysis, we found no protein in the human body like these three target proteins.
Cellular Location of Target Proteins
Prediction of CELLO2GO [30] reflects that Helicase is an extracellular protein. In case of, it may be extracellular or in the plasma membrane and the host cytoplasm. Furthermore, the PLpro protein position is in the plasma membrane and the cytoplasm of a host. Therefore, these drugs may interact easily for being extracellular [31-42].
Discussion
In emerging viral diseases like COVID-19, medical treatment usually starts with approved antiviral compounds, such as an FDA-approved polymerase inhibitor. Here, we tried to explore new antiviral compounds capable of inhibiting multiple essential proteins of the virus. Our study focused on (in silico) the identification of components that can interfere with the life cycle of SARS-CoV-2 through interacting with their non-structural proteins (nsp) instead of structural ones. Non-structural proteins such as RNA-dependent RNA polymerase (RdRp), Helicase (NSP13), and Papain Like protease (PLpro) have less sequence variability (Figure 1), solely involved in genome replication and the packaging of the virus into the host cell. Among them, Helicase is highly conserved in Coronaviridae family members.
Multiple Sequence Alignment (MSA) suggests that less than 1% amino acid mutation is present among (PLpro 0.05%, RdRp 0.2%, Helicase 0.16%) (Figure 1) these nsp in regional variants. Therefore, antiviral drugs targeting nsp’s may protect against SARS-CoV-2 irrespective of regional variants. This conservative nature of nsp (RdRp, Helicase, PLpro) suggests the possibility of long-term protection using drugs.
We performed homology modeling of these sole proteins in conjunction with the SWISS-MODEL database, and the outputs were saved in protein database (.pdb) format. ProtParam tool, known as ExPASy, is a database to determine a protein model’s physicochemical properties (Table 1). ExPASy suggests instability index values for proteins such as 8.12 for RdRp, 33.31 for Helicase, and 36.56 for L protease (Table 1). This index determines the stability of the protein model. Values higher than 40 indicate the instability of the model. As instability index values of three proteins are less than 40, structures of these proteins are highly stable.
The hydrophilicity or hydrophobicity of sole proteins was determined with a GRAVY (Grand Average of Hydrophobicity) score. GRAVY score below zero indicates hydrophobic and globular; whereas, a score over zero indicates hydrophilic. GRAVY scores of RdRp, PL Pro, Helicase are -0.218, -0.175, and -0.096, respectively (Table 1), meaning all three are hydrophobic, globular. In addition, the theoretical isoelectric (pI) points of PL protease, RdRp, and Helicase are 5.56, 6.14, and 8.66, respectively (Table 1). The pI of PL Protease (PLpro) speculates its acidic nature and Helicase slightly alkaline nature. Physicochemical properties show protein’s compatibility of making hydrogen bonds promptly.
The PROCHECK (Ramachandran Plotting), SAVES-Verify- 3D, PROSA databases were used for in-silico validation of protein models. Ramachandran Plotting showed RdRp, PL protease, and Helicase has 95.1%, 99.6%, 99% of amino acid residues in allowed regions, including 80.3%, 95%, and 76.3% are at the most favored area, respectively which assures high acceptability of protein models. SAVES-Verify-3D suggests that the proteins have valid 3D structures as they passed with a score over 80% (RdRp 83.72%, PLpro 89.14%, and Helicase 85.57%) (Table S1). PROSA, a web-based tool used to determine the overall quality of the protein models. It provides the folding energy of the 3D structure by the score of Z. Z-score of RdRp, Helicase, and PL protease is -8.9, -8.89, and -8.85, respectively (Figure 2). Moreover, RMSD values of these protein models with their respective template structure is under 2Å (Angstrom), which means the validity of models. These results speculate that the structures are native. Negative and values above eight indicate the stability of folding. Together, it reflects the validity of the protein models.
Drug Bank server suggested eight drugs (Table 2), including Remdesivir General, which can act against all three enzymes with variable binding efficiency (Table 2). Remdesivir General is now an approved medication to treat COVID-19. However, Remdesivir General has limitations, such as the low half-life in plasma (0.4h). Moreover, 60% of the Remdesivir General medicated patients faced adverse effects, such as renal impairment, hypotension, multiple organ dysfunction, increased hepatic enzyme, diarrhea to some extent (12%), septic shock, as well as kidney injury (approximately 10%). And 66% adverse events in adults, along with a 1.23 hazard ratio. DrugBank also suggests another drug that reacts only against PLpro protein named 2-(N-morpholino) ethanesulfonic acid (Table 2).
AutoDock Vina tool was used for molecular docking to screen and identify drug-like components with their binding efficacy with specified proteins. Molecular docking narrowed the choice of drug components into two, such Nalpha-[(benzyloxy) carbonyl]-N-[(1R)- 4-hydroxy-1-methyl-2-oxobutyl]-L-phenylalaninamide (DB08732), which have binding energy against RdRp - 13 Kcal/mole along with three alkyl bonds with the amino acid residues, Helicase – 12.5 Kcal/ mole with three alkyl and two other attractive charge bonds (Figure 4), and PLpro – 11.8 Kcal/mole (two alkyl bonds along with two other bonds) (Table 3). And, S- [5-( trifluoromethyl L)-4H-1,2,4- triazol-3-YL] 5-(phenylethyl) furan-2-carbothiote (DB07743), which showed binding energy against RdRp – 12.5 Kcal/mole (four alkyl and one halogen bond), Helicase – 13.4 Kcal/mole (3 Alkyl and 4 other bonds) (Figure 4), and PLpro – 11.4 Kcal/mole (Five Alkyl and three other bonds) (Table 3). These set of results reflect strong bonds along with higher binding energy support the effectiveness of drug like components (In silico). Furthermore, their ADMET profiles represented no carcinogenicity and had intestinal absorption capability with less toxicity and less corrosive. Moreover, pBLAST suggests that RdRp, Helicase (NSP13), and Papain-like protease (PLpro) have non-homologous characteristics with the proteins present in the Homo sapiens. As a result, these drug-like components will not interfere with any essential cell components. Thus, no harmful reaction will occur by these components. Therefore, both of them can be an excellent choice to incorporate into the drug discovery process.
In the future, in vitro experiments with these two components in a cell-line-based bioassay with live virus particle or pseudovirus will be very interesting to observe. In addition, further research on drug discovery prospects, coupled with in-vivo experiments, might provide us an alternative to vaccines.
To tackle the current pandemic and any future upsurge by RNA viruses like SARS-CoV-2, we, preemptively, need to develop antiviral drugs that can act irrespective of regional variants. Meanwhile, the vaccination program is ongoing worldwide, including Bangladesh, but different regional variants coupled with mutations in the spike protein remain a matter of concern. Thus, this study explored the possibility and importance of drug-like compounds as antiviral drugs to fight against the pandemic as a supportive drug and backup option for vaccine unavailability or failure.
Acknowledgement
This research study was funded by core donors who provide unrestricted support to icddr,b for its operations and research. Current donors providing unrestricted support include the Governments of Bangladesh, Canada, Sweden and the UK. We gratefully acknowledge our core donors for their support and commitment to icddr,b’s research efforts.
Declaration of Interest
The authors confirm that there are no known conflicts of interest associated with this publication.
References
- Ibrahim IM (2020) COVID-19 spike-host cell receptor GRP78 binding site prediction. J Infect 80: 554???562
- Chan (2015) Middle East respiratory syndrome coronavirus: another zoonotic betacoronavirus causing SARS-like disease. Clin Microbiol Rev 28:465-522
- Al-Qahtani AA (2020) Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2): emergence. History, Basic and Clinical Aspects. Saudi J Biol Sci 27: 2531???2538.
- Ren LL (2020) Identification of a novel coronavirus causing severe pneumonia in human: a descriptive study. Chinese Medical Journal 133: 1015-1024
- Marra MA (2003)The genome sequence of the SARS-associated coronavirus.SCIENCE 300:1399-1404
- Woo PC (2010) Coronavirus genomics and bioinformatics analysis. Viruses 2: 1804???1820
- Woo PC (2005) In silico analysis of ORF1ab in coronavirus HKU1 genome reveals a unique putative cleavage site of coronavirus HKU1 3C-like protease. Microbiol Immunol 49: 899???908
- Roose K, Fiers W, Saelens X (2009) Pandemic preparedness: toward a universal influenza vaccine. Drug News Perspect 22:80-92
- V???kovski P (2020) Coronavirus biology and replication: implications for SARS-CoV-2. Nat Rev Microbiol 28: 1???16
- Poduri RG Joshi, Jagadeesh G (2020) Drugs targeting various stages of the SARS-CoV-2 life cycle: exploring promising drugs for the treatment of Covid-19. Cell Signal 74: 109721-109721
- Thompson JD, Higgins DG, Gibson TJ (1994) CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice Nucleic Acids Res 22: 4673???4680
- Waterhouse A (2018) SWISS-MODEL: homology modelling of protein structures and complexes NuclAcid Res 46: W296???W303
- DeLano WL (2002) The PyMOL molecular graphics system. Scientific Research Publishing
- Laskowski RA (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Applied Crstyllography 283-291
- Eisenberg DR, L�thy, Bowie aJU (1997) VERIFY3D: assessment of protein models with three-dimensional profiles 396-404.
- Wiederstein M, Sippl MJ (2007) ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Research 35: W407???W410
- Gasteiger E (2005) Protein identification and analysis tools on the ExPASy server. The Proteomics Protocols Handbook 571-607.
- Ionescu CM (2015) Atomic Charge Calculator: interactive web-based calculation of atomic charges in large biomolecular complexes and drug-like molecules. Journal of Cheminformatics 1-13
- Wishart D (2018) Drug Bank 5.0: a major update to the Drug Bank database for 2018. Nucleic Acids Research 46: D1074???D1082
- Cheng F (2012) Admet SAR: a comprehensive source and free tool for assessment of chemical ADMET properties. ACS Publications 52: 3099???3105
- Trott O, Olson AJ (2010) AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 31:455-461
- O'Boyle NM (2011) Open Babel: An open chemical toolbox 33
- Studio D (2008) Discovery Studio.
- Boratyn GM (2013) BLAST: a more efficient report with usability improvements. Nucleic Acids Res 41: W29-W33
- Adedeji AO (2012) Mechanism of nucleic acid unwinding by SARS-CoV helicase.PLoS ONE 7: 3652- 36521
- Jo S (2020) Inhibition of SARS-CoV 3CL protease by flavonoids. J Enzyme Inhib Med Chem 35: 145-151
- Al-Horani RA, Kar S (2020) Potential Anti-SARS-CoV-2 Therapeutics That Target the Post-Entry Stages of the Viral Life Cycle: A Comprehensive Review 12: 1092
- Mahase E (2020) Covid-19: US approves remdesivir despite WHO trial showing lack of efficacy. British Medical Journal Publishing Group.
- Chakrabarty RP (2020) Identification and qualitative characterization of new therapeutic targets in Stenotrophomonas maltophilia through in silico proteome exploration. Microb Pathog 149: 104293
- Yu CS (2014) CELLO2GO: a web server for protein subCELlular Localization prediction with functional gene ontology annotation. Plos one 99368
- Aftab SO (2020) Analysis of SARS-CoV-2 RNA-dependent RNA polymerase as a potential therapeutic drug target using a computational approach. J Transl Med 18: 275
- Machitani M (2020) RNA-dependent RNA polymerase, RdRP, a promising therapeutic target for cancer and potentially COVID-19. Cancer Sci 11: 3976???3984
- Mirza MU, Froeyen M (2020) Structural elucidation of SARS-CoV-2 vital proteins: Computational methods reveal potential drug candidates against main protease, Nsp12 polymerase and Nsp13 helicase. J Pharm Anal 10: 320-328
- Gurung AB (2020) In silico structure modelling of SARS-CoV-2 Nsp13 helicase and Nsp14 and repurposing of FDA approved antiviral drugs as dual inhibitors. Gene Rep 21: 100860
- Dwivedi VDS, Arora, and Pandey A (2013) Computational analysis of physico-chemical properties and homology modeling of carbonic anhydrase from Cordyceps militaris 209-212
- Magdeldin S (2012) Murine colon proteome and characterization of the protein pathways 1-14
- Sarwar MW (2019) Homology modeling and docking analysis of ??C1 protein encoded by Cotton leaf curl Multan betasatellite with different plant flavonoids. Heliyon 5: e01303
- Xia X (2007) Protein isoelectric point 207-219.
- Rubin D (2020) FDA approval of remdesivir???a step in the right direction 2598-2600
- Hendaus MA (2020) Remdesivir in the treatment of Coronavirus Disease 2019 (COVID-19): A simplified summary 1-6
- Wang Y (2020) Remdesivir in adults with severe COVID-19: a randomised, double-blind, placebo-controlled. Multicenter. 395: 1569-1578
- Seeliger D, De Groot BL (2010) Ligand docking and binding site analysis with PyMOL and Autodock/Vina. Journal of Computer-Aided Molecular Design 24: 417???422.
| Name of the drug | Drug Bank ID | Mode of action |
|---|---|---|
| 2-(N-morpholino) ethane sulfonic acid | DB03814 | Unknown |
| Benzyl (2-oxopropyl) carbamate | DB07293 | Unknown |
| 2- [(2,4-Dichloro-5-methyl phenyl) sulfonyl]-1,3-dinitro-5-(trifluoromethyl)benzene | DB07620 | Unknown |
| S- [5-( trifluoromethyl L)-4H-1,2,4-triazol-3-YL] 5-(phenylethyl) furan-2-carbothiote | DB07743 | Unknown |
| 5-amino-2-methyl-N-[(1R)-1-naphthalen-1-ylethyl] benzamide | DB08656 | Unknown |
| Nalpha-[(benzyloxy) carbonyl]-N-[(1R)-4-hydroxy-1-methyl-2-oxobutyl]-L-phenylalaninamide | DB08732 | Unknown |
| 4-(Dimethylamino)benzoic acid | DB08748 | Unknown |
| Remdesivir General | DB14761 | Inhibitor |
| GS-441524 | DB15686 | Inhibitor |
Table S1: Pharmacological Mode Of Action Of Anti-SARS-Cov-2 Drug-Like Components.